Chapter 8 Manipulating Data
Like any analyst, I spend a lot of time manipulating the data to shape it into a desired format. Some say 90% of time spent on any data analysis is spent on data gathering and manipulation. R shines in this category. The dplyr, readr, and tidyr libraries make data manipulation easy.
Before we jump into the actual manipulation of data, let’s understand how dplyr works.
Let’s load the data first.
library(readr)
donor_data <- read_csv("data/DonorSampleDataCleaned.csv")dplyr offers many helpful functions to make data analysis easy. The following are some functions that I most commonly use.
count: This function automatically groups the categories and returns the number of rows in each category. For example, let’s say we want to find out the number of rows in our data for each gender type. We can do so with the following simple command.
library(dplyr)
count(donor_data, GENDER)
#> # A tibble: 6 x 2
#> GENDER n
#> <chr> <int>
#> 1 Female 16678
#> 2 Male 16233
#> 3 U 1
#> 4 Uknown 1091
#> 5 Unknown 12
#> 6 <NA> 493We can see that we need to clean up the gender column by combining the unknown values. This is very similar to the GROUP BY clause in SQL. The count function wraps the dplyr function group_by to only return the number of rows.
group_by: You can use this function to perform operations on a subset of data grouped by other columns. We can reproduce the previouscountexample usinggroup_byand a summary function calledtallyexplicitly.
tally(group_by(donor_data, GENDER))
#> # A tibble: 6 x 2
#> GENDER n
#> <chr> <int>
#> 1 Female 16678
#> 2 Male 16233
#> 3 U 1
#> 4 Uknown 1091
#> 5 Unknown 12
#> 6 <NA> 493summarize: Now that you can group the subsets of data using thegroup_byfunction, you can calculate any aggregate values for your analysis. Let’s say you want to find out average and median giving as well as average and median age by gender. The following is how to do so.
summarize(group_by(donor_data, GENDER),
avg_giving = mean(TotalGiving),
med_giving = median(TotalGiving),
avg_age = mean(AGE),
med_age = median(AGE))
#> # A tibble: 6 x 5
#> GENDER avg_giving med_giving avg_age med_age
#> <chr> <dbl> <dbl> <dbl> <int>
#> 1 Female 3282.6 25 NA NA
#> 2 Male 1590.1 25 NA NA
#> 3 U 0.0 0 41 41
#> 4 Uknown 788.0 15 NA NA
#> 5 Unknown 86.8 20 NA NA
#> 6 <NA> 286.4 20 NA NAYou will see that the age summary columns show “NA” values. That’s because R doesn’t remove NA or unknown or NULL values from aggregation. We need to remove those from the calculations by adding the na.rm = TRUE argument to the summarize function.
summarize(group_by(donor_data, GENDER),
avg_giving = mean(TotalGiving, na.rm = TRUE),
med_giving = median(TotalGiving, na.rm = TRUE),
avg_age = mean(AGE, na.rm = TRUE),
med_age = median(AGE, na.rm = TRUE))
#> # A tibble: 6 x 5
#> GENDER avg_giving med_giving avg_age med_age
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Female 3282.6 25 43.3 39
#> 2 Male 1590.1 25 48.1 46
#> 3 U 0.0 0 41.0 41
#> 4 Uknown 788.0 15 48.4 49
#> 5 Unknown 86.8 20 47.9 53
#> 6 <NA> 286.4 20 44.5 44mutate: This function adds a column to the provided data frame. This is very convenient when you want to perform some operations and add the result as a column to your data frame. For example, let’s say you want to convert theTotalGivingcolumn to its natural logarithmic value.
d1 <- mutate(donor_data, log_totalgiving = log(TotalGiving))
summary(d1$log_totalgiving)
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> -Inf -Inf 3.22 -Inf 4.97 16.32arrange: This function orders the data by the values in the given column. This is similar to theORDER BYclause inSQL. Let’s say you want to order the data frame by total giving. The following is one way to do so.
arrange(donor_data, TotalGiving)If you want to order the data in descending order, you need to add the desc function.
arrange(donor_data, desc(TotalGiving))select: This is exactly like theSELECTclause inSQL. This function will select only the columns you choose and return that subset of the data. You can also rename the columns while selecting.
For example, let’s say you want to select only TotalGiving and also want to rename it to TOTAL_GIVING. You can do so like this.
select(donor_data, TOTAL_GIVING = TotalGiving)
#> # A tibble: 34,508 x 1
#> TOTAL_GIVING
#> <dbl>
#> 1 10
#> 2 2100
#> 3 200
#> 4 0
#> 5 505
#> 6 0
#> # ... with 3.45e+04 more rowsfilter: This function is similar to theWHEREclause inSQL. You can filter the data by providing various criteria. For example, let’s say you want the subset of donors who gave more than $10,000 and have a valid email address.
filter(donor_data, TotalGiving > 10000 &
EMAIL_PRESENT_IND == 'Y')
#> # A tibble: 117 x 23
#> ID ZIPCODE AGE MARITAL_STATUS GENDER
#> <int> <chr> <int> <chr> <chr>
#> 1 72 47708 83 Married Male
#> 2 187 06359 32 <NA> Female
#> 3 416 46604 NA <NA> Male
#> 4 619 90265 59 <NA> Female
#> 5 782 39479 54 <NA> Female
#> 6 1046 99019 NA <NA> Male
#> # ... with 111 more rows, and 18 more variables:
#> # MEMBERSHIP_IND <chr>, ALUMNUS_IND <chr>,
#> # PARENT_IND <chr>, HAS_INVOLVEMENT_IND <chr>,
#> # WEALTH_RATING <chr>, DEGREE_LEVEL <chr>,
#> # PREF_ADDRESS_TYPE <chr>,
#> # EMAIL_PRESENT_IND <chr>, CON_YEARS <int>,
#> # PrevFYGiving <chr>, PrevFY1Giving <chr>,
#> # PrevFY2Giving <chr>, PrevFY3Giving <chr>,
#> # PrevFY4Giving <chr>, CurrFYGiving <chr>,
#> # TotalGiving <dbl>, DONOR_IND <chr>,
#> # BIRTH_DATE <date>==) in the code to compare the column values. You will get an error if you use a single equals (=) sign, which is reserved for assigning values.
8.1 Chaining Operations
Many of the dplyr functions are useful by themselves, but the power of dplyr is in chaining various functions together. Just like how you can extend a chain by adding a link, you can continue to operate on a data set by adding functions. This will be easy to see with an example. Let’s say you want to find out the number of donors, who have given over $10,000 and have an email address. You will use the “%>%” symbol to continue the chain.
donor_data %>% filter(TotalGiving > 10000 &
EMAIL_PRESENT_IND == 'Y') %>%
tally()
#> # A tibble: 1 x 1
#> n
#> <int>
#> 1 117But then you think you’d like to see the distribution of donors by gender. Easy-peasy.
donor_data %>% filter(TotalGiving > 10000 &
EMAIL_PRESENT_IND == 'Y') %>%
count(GENDER)
#> # A tibble: 4 x 2
#> GENDER n
#> <chr> <int>
#> 1 Female 58
#> 2 Male 56
#> 3 Uknown 2
#> 4 <NA> 1How about calculating the aggregates on these donors?
donor_data %>%
filter(TotalGiving > 10000 &
EMAIL_PRESENT_IND == 'Y') %>%
group_by(GENDER) %>%
summarize(avg_giving = mean(TotalGiving, na.rm = TRUE),
med_giving = median(TotalGiving, na.rm = TRUE),
avg_age = mean(AGE, na.rm = TRUE),
med_age = median(AGE, na.rm = TRUE))
#> # A tibble: 4 x 5
#> GENDER avg_giving med_giving avg_age med_age
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 Female 49303 17260 42.8 40
#> 2 Male 113929 23412 56.0 55
#> 3 Uknown 57354 57354 58.0 58
#> 4 <NA> 40788 40788 NaN NALet’s see further examples of data manipulation.
8.2 Selecting Columns
Let’s say that you want to create a new data set with a few columns from existing data. Say you want to select the IDs, wealth ratings, and total giving. You also want to rename the ID column to PROSPECT_ID and the TotalGiving column to TOTAL_GIVING to help your readers. You’d start with this.
donor_data %>%
select(PROSPECT_ID = ID,
WEALTH_RATING,
TOTAL_GIVING = TotalGiving)To see the changes, you can chain the glimpse function.
donor_data %>%
select(PROSPECT_ID = ID,
WEALTH_RATING,
TOTAL_GIVING = TotalGiving) %>%
glimpse()
#> Observations: 34,508
#> Variables: 3
#> $ PROSPECT_ID <int> 1, 2, 3, 4, 5, 6, 7, 8,...
#> $ WEALTH_RATING <chr> NA, NA, NA, NA, NA, NA,...
#> $ TOTAL_GIVING <dbl> 10, 2100, 200, 0, 505, ...But you need to save the results in a different data set.
donor_data_new <- donor_data %>%
select(PROSPECT_ID = ID,
WEALTH_RATING,
TOTAL_GIVING = TotalGiving)You can also reorder columns by changing the order of the columns listed in the select function.
donor_data %>%
select(PROSPECT_ID = ID,
TOTAL_GIVING = TotalGiving, WEALTH_RATING) %>%
glimpse()
#> Observations: 34,508
#> Variables: 3
#> $ PROSPECT_ID <int> 1, 2, 3, 4, 5, 6, 7, 8,...
#> $ TOTAL_GIVING <dbl> 10, 2100, 200, 0, 505, ...
#> $ WEALTH_RATING <chr> NA, NA, NA, NA, NA, NA,...You can also deselect a column by using a minus operation.
donor_data %>%
select(PROSPECT_ID = ID,
TOTAL_GIVING = TotalGiving,
-WEALTH_RATING) %>%
glimpse()
#> Observations: 34,508
#> Variables: 2
#> $ PROSPECT_ID <int> 1, 2, 3, 4, 5, 6, 7, 8, ...
#> $ TOTAL_GIVING <dbl> 10, 2100, 200, 0, 505, 0...If you want to move the ID column to the end, you could use the everything function.
donor_data %>%
select(-ID, everything(), ID) %>%
glimpse()
#> Observations: 34,508
#> Variables: 23
#> $ ZIPCODE <chr> "23187", "77643",...
#> $ AGE <int> NA, 33, NA, 31, 6...
#> $ MARITAL_STATUS <chr> "Married", NA, "M...
#> $ GENDER <chr> "Female", "Female...
#> $ MEMBERSHIP_IND <chr> "N", "N", "N", "N...
#> $ ALUMNUS_IND <chr> "N", "Y", "N", "Y...
#> $ PARENT_IND <chr> "N", "N", "N", "N...
#> $ HAS_INVOLVEMENT_IND <chr> "N", "Y", "N", "Y...
#> $ WEALTH_RATING <chr> NA, NA, NA, NA, N...
#> $ DEGREE_LEVEL <chr> NA, "UB", NA, NA,...
#> $ PREF_ADDRESS_TYPE <chr> "HOME", NA, "HOME...
#> $ EMAIL_PRESENT_IND <chr> "N", "Y", "N", "Y...
#> $ CON_YEARS <int> 1, 0, 1, 0, 0, 0,...
#> $ PrevFYGiving <chr> "$0", "$0", "$0",...
#> $ PrevFY1Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY2Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY3Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY4Giving <chr> "$0", "$0", "$0",...
#> $ CurrFYGiving <chr> "$0", "$0", "$200...
#> $ TotalGiving <dbl> 10, 2100, 200, 0,...
#> $ DONOR_IND <chr> "Y", "Y", "Y", "N...
#> $ BIRTH_DATE <date> NA, 1985-06-16, ...
#> $ ID <int> 1, 2, 3, 4, 5, 6,...select can perform renaming and selecting of columns at the same time. But if you just want to rename certain columns and still keep the rest of the data, you should use select’s cousin function, rename.
donor_data %>%
rename(PROSPECT_ID = ID, TOTAL_GIVING = TotalGiving) %>%
glimpse()
#> Observations: 34,508
#> Variables: 23
#> $ PROSPECT_ID <int> 1, 2, 3, 4, 5, 6,...
#> $ ZIPCODE <chr> "23187", "77643",...
#> $ AGE <int> NA, 33, NA, 31, 6...
#> $ MARITAL_STATUS <chr> "Married", NA, "M...
#> $ GENDER <chr> "Female", "Female...
#> $ MEMBERSHIP_IND <chr> "N", "N", "N", "N...
#> $ ALUMNUS_IND <chr> "N", "Y", "N", "Y...
#> $ PARENT_IND <chr> "N", "N", "N", "N...
#> $ HAS_INVOLVEMENT_IND <chr> "N", "Y", "N", "Y...
#> $ WEALTH_RATING <chr> NA, NA, NA, NA, N...
#> $ DEGREE_LEVEL <chr> NA, "UB", NA, NA,...
#> $ PREF_ADDRESS_TYPE <chr> "HOME", NA, "HOME...
#> $ EMAIL_PRESENT_IND <chr> "N", "Y", "N", "Y...
#> $ CON_YEARS <int> 1, 0, 1, 0, 0, 0,...
#> $ PrevFYGiving <chr> "$0", "$0", "$0",...
#> $ PrevFY1Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY2Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY3Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY4Giving <chr> "$0", "$0", "$0",...
#> $ CurrFYGiving <chr> "$0", "$0", "$200...
#> $ TOTAL_GIVING <dbl> 10, 2100, 200, 0,...
#> $ DONOR_IND <chr> "Y", "Y", "Y", "N...
#> $ BIRTH_DATE <date> NA, 1985-06-16, ...dplyr also offers various helper functions to select columns by conditions. Run ?select_helpers in your console to see all the functions.
The following are a few examples.
- Select all indicator columns.
donor_data %>%
select(ends_with('IND')) %>%
glimpse()
#> Observations: 34,508
#> Variables: 6
#> $ MEMBERSHIP_IND <chr> "N", "N", "N", "N...
#> $ ALUMNUS_IND <chr> "N", "Y", "N", "Y...
#> $ PARENT_IND <chr> "N", "N", "N", "N...
#> $ HAS_INVOLVEMENT_IND <chr> "N", "Y", "N", "Y...
#> $ EMAIL_PRESENT_IND <chr> "N", "Y", "N", "Y...
#> $ DONOR_IND <chr> "Y", "Y", "Y", "N...- Select all columns containing
Giving.
donor_data %>%
select(contains('Giving')) %>%
glimpse()
#> Observations: 34,508
#> Variables: 7
#> $ PrevFYGiving <chr> "$0", "$0", "$0", "$0",...
#> $ PrevFY1Giving <chr> "$0", "$0", "$0", "$0",...
#> $ PrevFY2Giving <chr> "$0", "$0", "$0", "$0",...
#> $ PrevFY3Giving <chr> "$0", "$0", "$0", "$0",...
#> $ PrevFY4Giving <chr> "$0", "$0", "$0", "$0",...
#> $ CurrFYGiving <chr> "$0", "$0", "$200", "$0...
#> $ TotalGiving <dbl> 10, 2100, 200, 0, 505, ...- Select all columns saved in a character vector.
select_these_cols <- c('DEGREE_LEVEL',
'PREF_ADDRESS_TYPE',
'ZIPCODE')
donor_data %>%
select(one_of(select_these_cols)) %>%
glimpse()
#> Observations: 34,508
#> Variables: 3
#> $ DEGREE_LEVEL <chr> NA, "UB", NA, NA, N...
#> $ PREF_ADDRESS_TYPE <chr> "HOME", NA, "HOME",...
#> $ ZIPCODE <chr> "23187", "77643", N...- Select columns 1 through 6.
donor_data %>%
select(1:6) %>% #can also be written as select(num_range())
glimpse()
#> Observations: 34,508
#> Variables: 6
#> $ ID <int> 1, 2, 3, 4, 5, 6, 7, 8...
#> $ ZIPCODE <chr> "23187", "77643", NA, ...
#> $ AGE <int> NA, 33, NA, 31, 68, 57...
#> $ MARITAL_STATUS <chr> "Married", NA, "Marrie...
#> $ GENDER <chr> "Female", "Female", "F...
#> $ MEMBERSHIP_IND <chr> "N", "N", "N", "N", "N...8.3 Filtering Rows
We’ve already learned how to filter rows, but let’s check out an example of chaining select and filter. Let’s say you want to select the IDs, wealth ratings, and total giving, but only those donors who gave more than $10,000 and have a valid email address.
donor_data %>%
filter(TotalGiving > 10000 &
EMAIL_PRESENT_IND == 'Y') %>%
select(PROSPECT_ID = ID,
WEALTH_RATING,
TOTAL_GIVING = TotalGiving) %>%
glimpse()
#> Observations: 117
#> Variables: 3
#> $ PROSPECT_ID <int> 72, 187, 416, 619, 782,...
#> $ WEALTH_RATING <chr> NA, NA, "$100,000-$249,...
#> $ TOTAL_GIVING <dbl> 125805, 17971, 20690, 1...The order of chaining is important for the operations to succeed. For example, if you use filter after select, what do you think will happen? Yes, exactly: The code won’t work because the TotalGiving and EMAIL_PRESENT_IND columns won’t exist after selecting only the ID, WEALTH_RATING, and TotalGiving columns.
##This code will give you 'object not found' error.
donor_data %>%
select(PROSPECT_ID = ID,
WEALTH_RATING,
TOTAL_GIVING = TotalGiving) %>%
#the data set now has only these columns:
# PROSPECT_ID, WEALTH_RATING, TOTAL_GIVING
filter(TotalGiving > 10000 &
EMAIL_PRESENT_IND == 'Y') %>%
#you can't filter what doesn't exist.
glimpse()8.4 Creating Columns
Often you will need to compute new values or add explanatory fields to your data sets. While select lets you rename fields, it doesn’t permit calculations on the fields. To do so, you can use the mutate function. Let’s say you want to add the natural log of value of the value of total giving as a column.
donor_data %>%
select(PROSPECT_ID = ID,
WEALTH_RATING,
TOTAL_GIVING = TotalGiving) %>%
mutate(TOTAL_GIVING_log = log(TOTAL_GIVING)) %>%
glimpse()
#> Observations: 34,508
#> Variables: 4
#> $ PROSPECT_ID <int> 1, 2, 3, 4, 5, 6, 7,...
#> $ WEALTH_RATING <chr> NA, NA, NA, NA, NA, ...
#> $ TOTAL_GIVING <dbl> 10, 2100, 200, 0, 50...
#> $ TOTAL_GIVING_log <dbl> 2.30, 7.65, 5.30, -I...You also want to create a column that shows whether the wealth rating is over $500,000. You can either modify the earlier mutate command or chain another mutate. For clarity’s sake, let’s add another mutate command.
But first, you want to see all the available values in the WEALTH_RATING column. You can either use the distinct function or just use `count. With a larger data set, finding unique values will be costlier on the CPU than summarizing.
donor_data %>%
count(WEALTH_RATING)
#> # A tibble: 9 x 2
#> WEALTH_RATING n
#> <chr> <int>
#> 1 $1-$24,999 580
#> 2 $1,000,000-$2,499,999 59
#> 3 $100,000-$249,999 511
#> 4 $2,500,000-$4,999,999 4
#> 5 $25,000-$49,999 564
#> 6 $250,000-$499,999 265
#> # ... with 3 more rowsdonor_data %>%
select(WEALTH_RATING) %>%
distinct(WEALTH_RATING)
#> # A tibble: 9 x 1
#> WEALTH_RATING
#> <chr>
#> 1 <NA>
#> 2 $50,000-$99,999
#> 3 $100,000-$249,999
#> 4 $25,000-$49,999
#> 5 $250,000-$499,999
#> 6 $1-$24,999
#> # ... with 3 more rowsWe know that to create our indicator column for the wealth rating over $500,000, we need to check for these wealth ratings: $500,000–$999,999; $1,000,000–$2,499,999; and $2,500,000–$4,999,999. We do so by placing these values in a vector and using ifelse and the “%in%” operator as follows.
donor_data %>%
select(PROSPECT_ID = ID,
WEALTH_RATING,
TOTAL_GIVING = TotalGiving) %>%
mutate(TOTAL_GIVING_log = log(TOTAL_GIVING)) %>%
mutate(IS_OVER_500K = ifelse(
WEALTH_RATING %in% c('$500,000-$999,999',
'$1,000,000-$2,499,999',
'$2,500,000-$4,999,999'),
'Yes', 'No')) %>%
glimpse()
#> Observations: 34,508
#> Variables: 5
#> $ PROSPECT_ID <int> 1, 2, 3, 4, 5, 6, 7,...
#> $ WEALTH_RATING <chr> NA, NA, NA, NA, NA, ...
#> $ TOTAL_GIVING <dbl> 10, 2100, 200, 0, 50...
#> $ TOTAL_GIVING_log <dbl> 2.30, 7.65, 5.30, -I...
#> $ IS_OVER_500K <chr> "No", "No", "No", "N...We can also get a quick count of prospects rated over $500,000.
donor_data %>%
select(PROSPECT_ID = ID,
WEALTH_RATING,
TOTAL_GIVING = TotalGiving) %>%
mutate(TOTAL_GIVING_log = log(TOTAL_GIVING)) %>%
mutate(IS_OVER_500K = ifelse(
WEALTH_RATING %in% c('$500,000-$999,999',
'$1,000,000-$2,499,999',
'$2,500,000-$4,999,999'),
'Yes', 'No')) %>%
count(IS_OVER_500K)
#> # A tibble: 2 x 2
#> IS_OVER_500K n
#> <chr> <int>
#> 1 No 34364
#> 2 Yes 1448.5 Creating Aggregate Data Frames
Let’s say that we want to calculate the giving statistics for alumni and non-alumni as well as the parent and non-parent population. How would we do so? We can group the data set by ALUMNUS_IND and PARENT_IND and then calculate the summary stats, like this.
alum_parent_summary <- donor_data %>%
group_by(ALUMNUS_IND, PARENT_IND) %>%
summarize(avg_giving = mean(TotalGiving, na.rm = TRUE),
med_giving = median(TotalGiving, na.rm = TRUE),
min_giving = min(TotalGiving, na.rm = TRUE),
max_giving = max(TotalGiving, na.rm = TRUE))
alum_parent_summary
#> # A tibble: 4 x 6
#> # Groups: ALUMNUS_IND [?]
#> ALUMNUS_IND PARENT_IND avg_giving med_giving
#> <chr> <chr> <dbl> <dbl>
#> 1 N N 1377 25
#> 2 N Y 1460 20
#> 3 Y N 5543 25
#> 4 Y Y 1529 35
#> # ... with 2 more variables: min_giving <dbl>,
#> # max_giving <dbl>8.6 Creating Joins
Aggregation is fun, but sometimes you need to combine your data with some other external data. For me, often, it is the ZIP Code-level data. The zipcode library provides the city, state, latitude, and longitude of each ZIP Code. So how do you integrate this data into your donor data? You use joins. The set operations dictate joins between two data sets. The most common data-manipulation and aggregation mistakes happen because of incorrect joins10. I recommend studying this topic in detail if you are not familiar with joins (here’s one resource for practitioners; and read Maier (1983) for theory.). But for completeness’ sake, here’s a quick overview.
Let’s say we have two data sets.
- Set A: This data contains the address and names of all our constituents, donors and non-donors
- Set B: This data contains total giving for every donor
Both the sets have a prospect ID, which we will use for joining. As donors are a subset of all of our constituents, set B is a subset of set A.
Here are some ways we can join set A and set B.
- Inner join: This join will return rows from both the sets only where the IDs are the same. That means all the non-donors will be excluded from the resulting set. The following is how you would write this in
SQL.
SELECT A.*, B.*
FROM A
INNER JOIN B on A.ID = B.ID- Outer join: Depending on which set is considered outer in the join, the returning result will contain all the rows from one set and only matching rows from the other set. For example, let’s say that we want to see all the constituents from set A along with their total giving. In this case, set B is the outer set. Here’s the
SQLcode for such a join.
SELECT A.*, B.*
FROM A
LEFT OUTER JOIN B on A.ID = B.IDSince set B is a subset of set A, the outer join between sets B and A will result in the same data as an inner join between them.
- Cross join: This join results in a set with the number of rows equal to the number of rows in the first set times the number of rows in the second set. This is a Cartesian product. This is the basis of joins, but we apply conditions to reduce the resulting set. The inner join is a Cartesian product with the criteria that the values of the two joining fields should match, resulting in a smaller set. Use cases of a cross join are limited and unless you have a good reason to use this type of join, find another way to get your results.
How do we use these joins, then? Let’s say that you want to get latitude and longitude values for each ZIP Code in your data. You can join your data with a zipcode data set. If we want the result set to contain only the ZIP Codes with a latitude and longitude in the zipcode data set, use an inner join. But if you want all the rows back from your original data set, use an outer join. Let’s see how to do both using dplyr.
library(zipcode)
data(zipcode)
glimpse(zipcode)
#> Observations: 44,336
#> Variables: 5
#> $ zip <chr> "00210", "00211", "00212", ...
#> $ city <chr> "Portsmouth", "Portsmouth",...
#> $ state <chr> "NH", "NH", "NH", "NH", "NH...
#> $ latitude <dbl> 43.0, 43.0, 43.0, 43.0, 43....
#> $ longitude <dbl> -71.0, -71.0, -71.0, -71.0,...
inner_join(donor_data, zipcode, by = c("ZIPCODE" = "zip")) %>%
glimpse()
#> Observations: 34,417
#> Variables: 27
#> $ ID <int> 1, 2, 4, 5, 6, 7,...
#> $ ZIPCODE <chr> "23187", "77643",...
#> $ AGE <int> NA, 33, 31, 68, 5...
#> $ MARITAL_STATUS <chr> "Married", NA, NA...
#> $ GENDER <chr> "Female", "Female...
#> $ MEMBERSHIP_IND <chr> "N", "N", "N", "N...
#> $ ALUMNUS_IND <chr> "N", "Y", "Y", "N...
#> $ PARENT_IND <chr> "N", "N", "N", "N...
#> $ HAS_INVOLVEMENT_IND <chr> "N", "Y", "Y", "N...
#> $ WEALTH_RATING <chr> NA, NA, NA, NA, N...
#> $ DEGREE_LEVEL <chr> NA, "UB", NA, NA,...
#> $ PREF_ADDRESS_TYPE <chr> "HOME", NA, "HOME...
#> $ EMAIL_PRESENT_IND <chr> "N", "Y", "Y", "Y...
#> $ CON_YEARS <int> 1, 0, 0, 0, 0, 3,...
#> $ PrevFYGiving <chr> "$0", "$0", "$0",...
#> $ PrevFY1Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY2Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY3Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY4Giving <chr> "$0", "$0", "$0",...
#> $ CurrFYGiving <chr> "$0", "$0", "$0",...
#> $ TotalGiving <dbl> 10, 2100, 0, 505,...
#> $ DONOR_IND <chr> "Y", "Y", "N", "Y...
#> $ BIRTH_DATE <date> NA, 1985-06-16, ...
#> $ city <chr> "Williamsburg", "...
#> $ state <chr> "VA", "TX", "IN",...
#> $ latitude <dbl> 37.3, 30.0, 38.5,...
#> $ longitude <dbl> -76.7, -93.9, -85...The vector used in the by argument, c("ZIPCODE" = "zip"), tells dplyr the columns to use for the join. If you didn’t want to see the city and state from the zipcode data, you could deselect those columns, like so.
inner_join(donor_data,
select(zipcode, -city, -state),
by = c("ZIPCODE" = "zip")) %>%
glimpse()
#> Observations: 34,417
#> Variables: 25
#> $ ID <int> 1, 2, 4, 5, 6, 7,...
#> $ ZIPCODE <chr> "23187", "77643",...
#> $ AGE <int> NA, 33, 31, 68, 5...
#> $ MARITAL_STATUS <chr> "Married", NA, NA...
#> $ GENDER <chr> "Female", "Female...
#> $ MEMBERSHIP_IND <chr> "N", "N", "N", "N...
#> $ ALUMNUS_IND <chr> "N", "Y", "Y", "N...
#> $ PARENT_IND <chr> "N", "N", "N", "N...
#> $ HAS_INVOLVEMENT_IND <chr> "N", "Y", "Y", "N...
#> $ WEALTH_RATING <chr> NA, NA, NA, NA, N...
#> $ DEGREE_LEVEL <chr> NA, "UB", NA, NA,...
#> $ PREF_ADDRESS_TYPE <chr> "HOME", NA, "HOME...
#> $ EMAIL_PRESENT_IND <chr> "N", "Y", "Y", "Y...
#> $ CON_YEARS <int> 1, 0, 0, 0, 0, 3,...
#> $ PrevFYGiving <chr> "$0", "$0", "$0",...
#> $ PrevFY1Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY2Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY3Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY4Giving <chr> "$0", "$0", "$0",...
#> $ CurrFYGiving <chr> "$0", "$0", "$0",...
#> $ TotalGiving <dbl> 10, 2100, 0, 505,...
#> $ DONOR_IND <chr> "Y", "Y", "N", "Y...
#> $ BIRTH_DATE <date> NA, 1985-06-16, ...
#> $ latitude <dbl> 37.3, 30.0, 38.5,...
#> $ longitude <dbl> -76.7, -93.9, -85...To see all the rows from the donor data, but only matching rows from the zipcode data, you can use the left outer join.
left_join(donor_data,
select(zipcode, -city, -state),
by = c("ZIPCODE" = "zip")) %>%
glimpse()
#> Observations: 34,508
#> Variables: 25
#> $ ID <int> 1, 2, 3, 4, 5, 6,...
#> $ ZIPCODE <chr> "23187", "77643",...
#> $ AGE <int> NA, 33, NA, 31, 6...
#> $ MARITAL_STATUS <chr> "Married", NA, "M...
#> $ GENDER <chr> "Female", "Female...
#> $ MEMBERSHIP_IND <chr> "N", "N", "N", "N...
#> $ ALUMNUS_IND <chr> "N", "Y", "N", "Y...
#> $ PARENT_IND <chr> "N", "N", "N", "N...
#> $ HAS_INVOLVEMENT_IND <chr> "N", "Y", "N", "Y...
#> $ WEALTH_RATING <chr> NA, NA, NA, NA, N...
#> $ DEGREE_LEVEL <chr> NA, "UB", NA, NA,...
#> $ PREF_ADDRESS_TYPE <chr> "HOME", NA, "HOME...
#> $ EMAIL_PRESENT_IND <chr> "N", "Y", "N", "Y...
#> $ CON_YEARS <int> 1, 0, 1, 0, 0, 0,...
#> $ PrevFYGiving <chr> "$0", "$0", "$0",...
#> $ PrevFY1Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY2Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY3Giving <chr> "$0", "$0", "$0",...
#> $ PrevFY4Giving <chr> "$0", "$0", "$0",...
#> $ CurrFYGiving <chr> "$0", "$0", "$200...
#> $ TotalGiving <dbl> 10, 2100, 200, 0,...
#> $ DONOR_IND <chr> "Y", "Y", "Y", "N...
#> $ BIRTH_DATE <date> NA, 1985-06-16, ...
#> $ latitude <dbl> 37.3, 30.0, NA, 3...
#> $ longitude <dbl> -76.7, -93.9, NA,...8.7 Transforming Data
Often you see the need to transform or pivot the data so that it becomes easier to perform further computations. You can do so in SQL using complicated CASE, PARTITION, or PIVOT statements. But the tidyr package makes the whole process easy in R.
Let’s consider two types of data layouts. The most typical data layout is the wide (a.k.a. flat) layout, which is one row per ID and all other descriptive information in columns as shown in Figure 8.1. The other common layout is the long layout, which is multiple rows per ID with different information in each row as shown in Figure 8.2. The donor sample file we have been using so far has the wide layout, and a typical gift transaction table has a long layout.
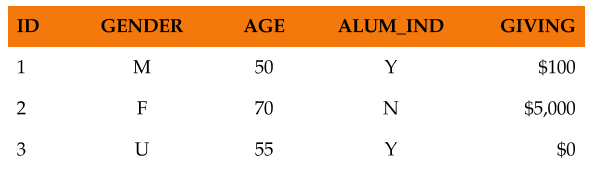
FIGURE 8.1: Wide layout

FIGURE 8.2: Long layout
8.7.1 Transforming from Wide to Long Layout
Let’s say you want to transform the donor data file so that all the indicator column types are in one column and the resulting values in another column. Let’s get started by loading the tidyr library.
library(tidyr)We will then use the gather function to combine the columns.
donor_data %>%
select(ID, ends_with("IND")) %>%
gather(key = "WHICH_IND", value = "IND_VALUE", -ID) %>%
#just to show different values in the resulting data set
sample_n(size = 15)
#> # A tibble: 15 x 3
#> ID WHICH_IND IND_VALUE
#> <int> <chr> <chr>
#> 1 4388 EMAIL_PRESENT_IND N
#> 2 32892 PARENT_IND N
#> 3 2439 PARENT_IND N
#> 4 33481 DONOR_IND Y
#> 5 5919 EMAIL_PRESENT_IND N
#> 6 2216 MEMBERSHIP_IND N
#> # ... with 9 more rowsWe provided the value of WHICH_IND to the key argument. This lets tidyr know to create a column named WHICH_IND housing all the different indicator column names. We also provided the value of IND_VALUE to the value argument. By doing this, tidyr created a column named IND_VALUE to house all the various values from the indicator columns. The last argument of -ID was to remove the ID column from the combination and instead use it as another column.
Another more meaningful example: Let’s say that you want to combine all the previous years’ giving columns together into one column.
donor_data %>%
select(ID, starts_with("PrevFY")) %>%
gather(key = "WHICH_FY", value = "GIVING", -ID) %>%
#just to show different values in the resulting data set
sample_n(size = 15)
#> # A tibble: 15 x 3
#> ID WHICH_FY GIVING
#> <int> <chr> <chr>
#> 1 17928 PrevFYGiving $0
#> 2 3159 PrevFY2Giving $0
#> 3 11649 PrevFY4Giving $0
#> 4 26287 PrevFY1Giving $0
#> 5 32751 PrevFY1Giving $0
#> 6 31136 PrevFY1Giving $0
#> # ... with 9 more rows8.7.2 Transforming Data from Long to Wide Layout
Let’s say that you want to transform the giving data file so that all the fiscal years are column names and the values under each column show the giving in that fiscal year. This is a typical pivot table example. For this example, we will use the RFM data file. Let’s load this file and see the contents.
giving_data <- read_csv("data/SampleDataRFM.csv")
glimpse(giving_data)
#> Observations: 6,973
#> Variables: 3
#> $ ID <int> 1, 1, 2, 2, 2, 2, 2, 3, 3...
#> $ FISCAL_YEAR <int> 2015, 2016, 2012, 2013, 2...
#> $ Giving <chr> "$1,000", "$600", "$100",...Now, let’s create new columns with the fiscal years as column names and fill them with the giving of that respective year and donor. We will use the spread function.
giving_data %>%
spread(key = FISCAL_YEAR, value = Giving, fill = 0) %>%
head()
#> # A tibble: 6 x 6
#> ID `2012` `2013` `2014` `2015` `2016`
#> <int> <chr> <chr> <chr> <chr> <chr>
#> 1 1 0 0 0 $1,000 $600
#> 2 2 $100 $55 $160 $135 $1,135
#> 3 3 $1,750 0 $1,065 $80 $35
#> 4 4 0 $555 0 $285 $885
#> 5 5 $1,000 $555 0 0 0
#> 6 7 $1,000 $250 0 0 0R doesn’t take column names that start with a number; therefore, tidyr automatically puts the column names in back ticks (```). This creates a problem when you want to select this type of column.
You can’t use this.
#don't run
giving_data %>%
spread(key = FISCAL_YEAR, value = Giving, fill = 0) %>%
select(2012)Because the select function expects a column at the “2012” position. You can select that column by surrounding it in back ticks. It becomes cumbersome quickly.
You have two options.
- Change the value of the column before applying
spread. I prefer this option because I know I need to modify only one column. - Rename the columns after applying
spread. This is not a bad option, but you do need to provide names for all the columns at their respective positions. This is not repeatable.
But let’s try both.
Changing the values of the column by prefixing the year values with FY.
giving_data %>%
mutate(FISCAL_YEAR = paste0("FY", FISCAL_YEAR)) %>%
spread(key = FISCAL_YEAR, value = Giving, fill = 0) %>%
head()
#> # A tibble: 6 x 6
#> ID FY2012 FY2013 FY2014 FY2015 FY2016
#> <int> <chr> <chr> <chr> <chr> <chr>
#> 1 1 0 0 0 $1,000 $600
#> 2 2 $100 $55 $160 $135 $1,135
#> 3 3 $1,750 0 $1,065 $80 $35
#> 4 4 0 $555 0 $285 $885
#> 5 5 $1,000 $555 0 0 0
#> 6 7 $1,000 $250 0 0 0Renaming the resulting columns.
giving_data %>%
spread(key = FISCAL_YEAR, value = Giving, fill = 0) %>%
setNames(., c('ID', 'FY2012', 'FY2013',
'FY2014', 'FY2015', 'FY2016')) %>%
head()
#> # A tibble: 6 x 6
#> ID FY2012 FY2013 FY2014 FY2015 FY2016
#> <int> <chr> <chr> <chr> <chr> <chr>
#> 1 1 0 0 0 $1,000 $600
#> 2 2 $100 $55 $160 $135 $1,135
#> 3 3 $1,750 0 $1,065 $80 $35
#> 4 4 0 $555 0 $285 $885
#> 5 5 $1,000 $555 0 0 0
#> 6 7 $1,000 $250 0 0 0rep function.
If you’re enjoying this book, consider sharing it with your network by running
source("http://arn.la/shareds4fr")in yourRconsole.— Ashutosh and Rodger
References
Maier, David. 1983. The Theory of Relational Databases. Vol. 11. Computer science press Rockville. http://web.cecs.pdx.edu/~maier/TheoryBook/TRD.html.
The joins by themselves are rarely incorrect; the mistakes happen because of poor understanding of what happens after joining two sets. If one data set has multiple rows for a field used for joining another data set, the resulting join will have multiple rows.↩