Chapter 12 Machine Learning Recipes
Machine learning is the study of computer-based algorithms to build models that learn and make predictions using data.
George Box, a noted statistician, is often credited for saying “all models are wrong” in his paper Science and Statistics (Box 1976). Box’s quote expanded to “all models are wrong, but some are useful” in the popular text Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach (Burnham 2002).
All models are wrong, but some are useful.
— George Box
Many machine learning tasks draw from pattern-recognition concepts in artificial intelligence (AI), which is a discipline focused on enabling computers to learn concepts and functions without explicit programming. In terms of real-world applications, machine learning is widely used to design and build models that can solve a variety of complex computing tasks and make useful predictions using data. Examples you might recognize include spam filtering, fraud detection, and optical character recognition (OCR).
12.1 Feature Selection
As you build your own models, it is important to first identify the specific types of problems you are trying to solve with your model and its outputs. It is also important to think about what kinds of data you will need to find, use, and test with your models.
Most of the recipes in this chapter focus on building models that predict whether a constituent is a donor or non-donor. By design, we are focusing on the same donor modeling problem so you can develop a sense of the similarities and differences between various machine learning methods. We also selected this modeling problem due to the fundamental and pressing need in non-profits to identify potential donors. Fortunately, many of you reading this book already have access to heaps of institutional data and, by following the recipes in this chapter, you will hopefully be inspired to take a fresh look at your data and modify these recipes to generate actionable insights.
Feature selection is the process of choosing input variables or relevant attributes for your model. To quickly introduce you to multiple machine learning methods, we opted to simplify this process and preselect some features (variables) from our test data set, such as age, constituent type, involvement indicators, and communication preferences.
The caret package includes tools that you can use to rank the importance of your data variables and automatically select the most important features (variables) to potentially incorporate into your models. One popular automatic method for feature selection provided by the caret package is Recursive Feature Elimination (RFE). To read more about using RFE, check out this tutorial.
In addition to the caret package, the mlr package also offers methods to calculate feature importance and guide your feature selection process. To read more about using mlr’s filtering and wrapper methods to identify the most relevant feature, check out this article.
12.2 Supervised Learning
The purpose of supervised learning is to make accurate predictions using labeled data.
Typically, there are two types of predictions: categorical (classification) and numeric (regression).
12.2.1 Classification
Classification, which is a type of supervised learning, uses labeled input (training) data to predict categorical (or discrete) values on new (test) data. The classification model is built with training data containing known categorical labels, which are used to predict the unknown category values of new (test) observations.
12.2.2 Instance-Based Learning
Instance-based learning is a machine learning method that constructs a hypothesis directly from an entire training dataset. Instance-based learning, also known as memory-based or lazy learning, is one of the simplest types of machine learning algorithms.
Instance-based learning models store training data, compute similarities (that is, distance measurements such as Euclidean, Manhattan, Canberra, and Minkowski) between training and test instances, and decide (predict) the value or class for a new observation of test data.
R, write the command ?dist into the R console.
The primary advantage of instance-based (or memory-based) models over other machine learning methods is the ability to instantly adapt learning models to new data. The key disadvantage is that instance-based learning methods require significant computational resources to store entire training datasets into memory all at once, which can present limitations when working with large datasets with many attributes or variables (also known as features). In addition, instance-based models are prone to overfitting noise (error) in training data, which is important to keep in mind as you dive further into building models and exploring various machine learning techniques.
12.2.2.1 K-Nearest Neighbor
The K-nearest neighbor (KNN) algorithm is a commonly used instance-based learning method and one of the simplest machine learning algorithms available.
The KNN algorithm can be used for both classification (categorical) and regression (numeric) prediction problems. In KNN classification, the output is a categorical (discrete) value such as gender, constituent type, or marital status. In KNN regression, the output is a numerical (continuous) value of an object attribute.
KNN is a non-parametric learning method, which means that it does not make any assumptions about your input data or its distribution. Because KNN stores its entire training dataset into memory, it does not require an explicit learning phase like other machine learning algorithms. Instead, KNN seeks to identify patterns within your input data (k closest observations) using a distance measurement (Euclidean, by default) and majority vote to classify (predict) each data point rather than fitting your data to a particular model or function.
Let’s begin preparing our data to build a KNN classification model:
# Load dplyr package
library(dplyr)
library(readr)
# Load string package
library(stringr)
# Load Data
donor_data <- read_csv("data/DonorSampleDataML.csv")Using the glimpse function from the dplyr library, we can see that the donor data file is structured with 34,508 examples (observations) and 23 features (variables).
# Glimpse donor_data
glimpse(donor_data)Before building our classification model, we should drop the “ID” variable since it is not appropriate to use as a classification feature.
# Drop 'ID' variable
donor_data <- select(donor_data, -ID)The DONOR_IND variable, which indicates Y or N, is the variable (feature) we seek to classify (predict) based on input (training) data. Let’s use the table function to count the distribution of donors and non-donors in the example data file.
# Table of Donor Count
table(donor_data$DONOR_IND)
#>
#> N Y
#> 13075 21433Based on the table output, there are 13,075 non-donors and 21,433 donors. Even though the majority (62%) of constituents are donors in this dataset, it is important to note that this is an artificially generated dataset and that your mileage (that is, donors per constituent record) will vary based on your institutional activity and transactional record.
Now let’s use the CrossTable function, from the gmodels library (Warnes et al. 2015), to produce a two-way contingency table (also known as cross-tabulation) that compares donor and parent constituent type indicators (Y or N) to identify the relative proportion of overall donor parents in the data file.
# Install gmodels package
install.packages("gmodels",
repos = "http://cran.us.r-project.org")# Load gmodels
library(gmodels)
# Use CrossTable
CrossTable(x = donor_data$ALUMNUS_IND,
y = donor_data$DONOR_IND,
chisq = FALSE)
#>
#>
#> Cell Contents
#> |-------------------------|
#> | N |
#> | Chi-square contribution |
#> | N / Row Total |
#> | N / Col Total |
#> | N / Table Total |
#> |-------------------------|
#>
#>
#> Total Observations in Table: 34508
#>
#>
#> | donor_data$DONOR_IND
#> donor_data$ALUMNUS_IND | N | Y | Row Total |
#> -----------------------|-----------|-----------|-----------|
#> N | 10800 | 297 | 11097 |
#> | 10345.493 | 6311.171 | |
#> | 0.973 | 0.027 | 0.322 |
#> | 0.826 | 0.014 | |
#> | 0.313 | 0.009 | |
#> -----------------------|-----------|-----------|-----------|
#> Y | 2275 | 21136 | 23411 |
#> | 4903.846 | 2991.545 | |
#> | 0.097 | 0.903 | 0.678 |
#> | 0.174 | 0.986 | |
#> | 0.066 | 0.612 | |
#> -----------------------|-----------|-----------|-----------|
#> Column Total | 13075 | 21433 | 34508 |
#> | 0.379 | 0.621 | |
#> -----------------------|-----------|-----------|-----------|
#>
#> Based on the relative proportions in the cross-table output, it appears that alumni (68%) tend to give much more frequently on average than their non-alumni (32%) counterparts. Specifically, the cross-table reveals that 90% of alumni give and 10% do not give in this example dataset.
Now, let’s explore how to use the KNN algorithm to build a classification model that predicts whether a constituent is a donor or non-donor using input (training) data features such as age, constituent type, involvement indicators, and communication preferences.
First let’s install the class library, which provides various classification functions (Ripley 2015). We will also use the caret library for model evaluation (Jed Wing et al. 2017).
# Install class package
install.packages("class",
repos = "http://cran.us.r-project.org")# Load dplyr
library(dplyr)
# Load class package
library(class)
# Load caret package
library(caret)
# Set Seed for Repeatable Results
set.seed(777)Since the function expects numeric columns, let’s convert some of the factors to numeric.
# Convert from character to numeric data type
convert_fac2num <- function(x){
as.numeric(as.factor(x))
}
# Convert features from factor to numeric
donor_data <- mutate_at(donor_data,
.vars = c('MARITAL_STATUS', 'GENDER',
'ALUMNUS_IND', 'PARENT_IND',
'HAS_INVOLVEMENT_IND', 'DEGREE_LEVEL',
'PREF_ADDRESS_TYPE', 'EMAIL_PRESENT_IND'),
.funs = convert_fac2num)
# Convert feature to factor
donor_data$DONOR_IND <- as.factor(donor_data$DONOR_IND)Next, let’s split 70% of the data into training data and 30% into test data.
dd_index <- sample(2, nrow(donor_data),
replace = TRUE,
prob = c(0.7, 0.3))
dd_trainset <- donor_data[dd_index == 1, ]
dd_testset <- donor_data[dd_index == 2, ]
# Confirm size of training and test
# datasets
dim(dd_trainset)
dim(dd_testset)
# Store class labels
dd_trainset_labels <- dd_trainset$DONOR_IND[1:5000]
dd_testset_labels <- dd_testset$DONOR_IND[1:5000]Since we need only numeric columns for this function, let’s select the numeric variables.
dd_trainset <- select_if(dd_trainset[1:5000, ], is.numeric)
dd_testset <- select_if(dd_testset[1:5000, ], is.numeric) We need to convert the numeric values to a range between 0 and 1. Let’s do so by defining a min-max normalization function.
min_max <- function(x) {
return((x - min(x))/(max(x) - min(x)))
}Next, let’s apply this function to all the columns.
dd_trainset_n <- mutate_all(dd_trainset_n,
min_max)
dd_testset_n <- mutate_all(dd_testset_n,
min_max)Now, let’s train multiple KNN models using different cluster sizes, where k specifies the size of each cluster.
dd_test_pred_k1 <- knn(train = dd_trainset_n,
test = dd_testset_n, cl = dd_trainset_labels[1:5000],
k = 1)
dd_test_pred_k5 <- knn(train = dd_trainset_n,
test = dd_testset_n, cl = dd_trainset_labels[1:5000],
k = 5)
dd_test_pred_k10 <- knn(train = dd_trainset_n,
test = dd_testset_n, cl = dd_trainset_labels[1:5000],
k = 10)
dd_test_pred_k32 <- knn(train = dd_trainset_n,
test = dd_testset_n, cl = dd_trainset_labels[1:5000],
k = 32)Let’s see how the models performed using the confusion matrix.
confusionMatrix(table(dd_test_pred_k1,
dd_testset_labels), positive = "Y")
confusionMatrix(table(dd_test_pred_k5,
dd_testset_labels), positive = "Y")
confusionMatrix(table(dd_test_pred_k10,
dd_testset_labels), positive = "Y")
confusionMatrix(table(dd_test_pred_k32,
dd_testset_labels), positive = "Y")Instead of normalizing all the values between 0 and 1, let’s scale all the values that are centered at 0 using the scale function.
# Apply z-score standardization to
# training and test datasets
dd_trainset_zscore <- as.data.frame(scale(dd_trainset))
dd_testset_zscore <- as.data.frame(scale(dd_testset))Let’s build models using these scaled values.
dd_test_pred_zs_k1 <- knn(
train = dd_trainset_zscore,
test = dd_testset_zscore,
cl = dd_trainset_labels[1:5000],
k = 1)
dd_test_pred_zs_k5 <- knn(
train = dd_trainset_zscore,
test = dd_testset_zscore,
cl = dd_trainset_labels[1:5000],
k = 5)
dd_test_pred_zs_k10 <- knn(
train = dd_trainset_zscore,
test = dd_testset_zscore,
cl = dd_trainset_labels[1:5000],
k = 10)
dd_test_pred_zs_k32 <- knn(
train = dd_trainset_zscore,
test = dd_testset_zscore,
cl = dd_trainset_labels[1:5000],
k = 32)Let’s see whether the scaling helped.
confusionMatrix(table(dd_test_pred_zs_k1,
dd_testset_labels), positive = "Y")
confusionMatrix(table(dd_test_pred_zs_k5,
dd_testset_labels), positive = "Y")
confusionMatrix(table(dd_test_pred_zs_k10,
dd_testset_labels), positive = "Y")
confusionMatrix(table(dd_test_pred_zs_k32,
dd_testset_labels), positive = "Y")Using the KNN algorithm, we were able to predict whether a constituent was a donor or non-donor with 92% accuracy using a k (neighbors) value of 10, which produced 141 false negatives and 263 false positives.
To improve the results of the KNN model, we alternatively used the scale function to rescale our numerical input data values into z-scores and built a second classifier with 93% accuracy using a k (neighbors) value of 10, which produced 120 false negatives and 239 false positives.
12.2.3 Probabilistic Machine Learning
Probabilistic machine learning uses statistical metrics to classify (predict) test values based on input (training) data about prior events.
12.2.3.1 Naive Bayes
Naive Bayes is one of the most commonly used machine learning methods for classification tasks such as spam filtering, and fraud detection
As suggested by its name, the naive Bayes algorithm makes a couple of “naive” assumptions about your input data. Specifically, naive Bayes assumes that all of the features in your data are independent and equally important. Despite these rather unrealistic set of assumptions, the naive Bayes algorithm is useful and performs relatively well in many kinds of real-world applications and contexts. Naive Bayes is an especially popular choice for text-classification learning problems, which we will further explore in the Text Mining chapter.
Since we changed the underlying data in the previous example, going forward we will reload the data and manipulate it so that the format works for the technique used in the following examples.
# Load readr
library(readr)
# Load dplyr
library(dplyr)
# Load e1071
library(e1071)
# Load caret package
library(caret)
# Set Seed for Repeatable Results
set.seed(123)
# Load Data
donor_data <- read_csv("data/DonorSampleDataML.csv")Let’s select the variables (features) we need for this example.
pred_vars <- c('MARITAL_STATUS', 'GENDER',
'ALUMNUS_IND', 'PARENT_IND',
'HAS_INVOLVEMENT_IND', 'DEGREE_LEVEL',
'PREF_ADDRESS_TYPE', 'EMAIL_PRESENT_IND',
'DONOR_IND')
donor_data <- select(donor_data,
pred_vars,
AGE)Next, let’s convert the character variables to factor type.
donor_data <- mutate_at(donor_data,
.vars = pred_vars,
.funs = as.factor)Similar to the example above, let’s split the data into test and training.
dd_index <- sample(2, nrow(donor_data),
replace = TRUE,
prob = c(0.7, 0.3))
dd_trainset <- donor_data[dd_index == 1, ]
dd_testset <- donor_data[dd_index == 2, ]
# Confirm size of training and test
# datasets
dim(dd_trainset)
#> [1] 24287 10
dim(dd_testset)
#> [1] 10221 10Now, the easy part. Let’s build the naive Bayes classification model from the e1071 library (Meyer et al. 2017).
dd_naivebayes <- naiveBayes(
DONOR_IND ~ MARITAL_STATUS + GENDER
+ ALUMNUS_IND + PARENT_IND + HAS_INVOLVEMENT_IND
+ DEGREE_LEVEL + EMAIL_PRESENT_IND + AGE,
data = dd_trainset)Let’s see how the model performed.
dd_naivebayes
#>
#> Naive Bayes Classifier for Discrete Predictors
#>
#> Call:
#> naiveBayes.default(x = X, y = Y, laplace = laplace)
#>
#> A-priori probabilities:
#> Y
#> N Y
#> 0.377 0.623
#>
#> Conditional probabilities:
#> MARITAL_STATUS
#> Y Divorced Married Never Married Separated
#> N 2.62e-03 1.74e-01 1.09e-04 1.09e-04
#> Y 2.05e-03 1.97e-01 6.61e-05 3.30e-04
#> MARITAL_STATUS
#> Y Single Unknown Widowed
#> N 1.00e-01 7.19e-01 4.15e-03
#> Y 8.70e-02 7.08e-01 5.62e-03
#>
#> GENDER
#> Y Female Male Unknown
#> N 0.4876 0.4600 0.0524
#> Y 0.4761 0.4796 0.0443
#>
#> ALUMNUS_IND
#> Y N Y
#> N 0.8276 0.1724
#> Y 0.0136 0.9864
#>
#> PARENT_IND
#> Y N Y
#> N 0.9825 0.0175
#> Y 0.9246 0.0754
#>
#> HAS_INVOLVEMENT_IND
#> Y N Y
#> N 0.782 0.218
#> Y 0.750 0.250
#>
#> DEGREE_LEVEL
#> Y Graduate Non-Alumni Undergrad
#> N 0.0000 0.8337 0.1663
#> Y 0.1130 0.0136 0.8734
#>
#> EMAIL_PRESENT_IND
#> Y N Y
#> N 0.687 0.313
#> Y 0.659 0.341
#>
#> AGE
#> Y [,1] [,2]
#> N 31.4 11.8
#> Y 43.7 11.2Let’s make predictions on the test data set using the predict function.
donor_prediction <- predict(
dd_naivebayes, select(dd_testset,
pred_vars,
AGE))Did the predictions work? Let’s check out the confusion matrix.
naivebayes.crosstab <- table(donor_prediction, dd_testset$DONOR_IND)
confusionMatrix(naivebayes.crosstab, positive = "Y")
#> Confusion Matrix and Statistics
#>
#>
#> donor_prediction N Y
#> N 3241 91
#> Y 677 6212
#>
#> Accuracy : 0.925
#> 95% CI : (0.92, 0.93)
#> No Information Rate : 0.617
#> P-Value [Acc > NIR] : <2e-16
#>
#> Kappa : 0.836
#> Mcnemar's Test P-Value : <2e-16
#>
#> Sensitivity : 0.986
#> Specificity : 0.827
#> Pos Pred Value : 0.902
#> Neg Pred Value : 0.973
#> Prevalence : 0.617
#> Detection Rate : 0.608
#> Detection Prevalence : 0.674
#> Balanced Accuracy : 0.906
#>
#> 'Positive' Class : Y
#> Using the naive Bayes algorithm, we predicted whether a constituent was a donor or non-donor with 92.49% accuracy, which produced 648 false negatives and 85 false positives.
laplace values in your naiveBayes() function. Can you increase the accuracy of the model using different laplace values? Write the ?naiveBayes command in your R console for additional documentation details.
We will explore how to use the naive Bayes machine learning method to build a text classifier, make predictions, and evaluate results in the Text Mining chapter.
For now, let’s continue exploring different types of machine learning algorithms to increase your overall exposure, familiarity, and understanding of various algorithms you will likely encounter and/or use in your data analytics journey.
12.2.4 Decision Trees
Decision trees are fundamental machine learning tools that help explore the structure of input (training) data by providing visual decision rules that predict classification (categorical) or regression (continuous) outcomes.
Decision trees (also known as tree-based models) use a split condition method to classify (predict) class or group membership based on one or more input variables. The classification process begins at the root node of the decision tree and repeatedly checks whether it should proceed to the right or left sub-branch node based on the split condition criteria, evaluating the input variable value at each node. The decision tree (also known as a recursive partitioning tree) is built once this recursive splitting process is completed.
For additional details about decision trees, you may refer to the Decision Tree Learning article.
12.2.4.1 CART
Classification and Regression Tree (also known as CART) models can be generated using the rpart package.
Suppose you are preparing for an upcoming fundraising campaign and need to identify, prioritize, and engage various constituents (for example, alumni, parents, and friends) to support your institution. Using example data and the rpart package (Therneau, Atkinson, and Ripley 2017), let’s train a classification model that predicts whether a constituent is a donor or non-donor based on available input (training) data such as demographic factors, educational level, involvement indicators, and communication preferences.
12.2.4.2 rpart
First, let’s install the rpart and rpart.plot libraries (Therneau, Atkinson, and Ripley 2017; Milborrow 2017).
# Install rpart.plot
install.packages(c("rpart", "rpart.plot"),
repos = 'http://cran.us.r-project.org')Next, let’s load the data, select model variables, and change the column types.
# Load readr
library(readr)
# Load rpart
library(rpart)
# Load rpart.plot
library(rpart.plot)
# Load caret library
library(caret)
# Load Data
donor_data <- read_csv("data/DonorSampleDataML.csv")
pred_vars <- c('MARITAL_STATUS', 'GENDER',
'ALUMNUS_IND', 'PARENT_IND',
'HAS_INVOLVEMENT_IND', 'DEGREE_LEVEL',
'PREF_ADDRESS_TYPE', 'EMAIL_PRESENT_IND',
'DONOR_IND')
donor_data <- select(donor_data,
pred_vars,
AGE)
# Set Seed for Repeatable Results
set.seed(777)
# Convert features to factor
donor_data <- mutate_at(donor_data,
.vars = pred_vars,
.funs = as.factor)Let’s divide the data into test and training data sets.
dd_index <- sample(2,
nrow(donor_data),
replace = TRUE,
prob = c(0.7, 0.3))
dd_trainset <- donor_data[dd_index == 1, ]
dd_testset <- donor_data[dd_index == 2, ]
# Confirm size of training and test
# datasets
dim(dd_trainset)
#> [1] 24195 10
dim(dd_testset)
#> [1] 10313 10Now, let’s build the decision tree using the rpart function.
dd.rp <- rpart(
DONOR_IND ~ .,
method = "class",
control = rpart.control(minsplit = 2,
minbucket = 2,
cp = 0.001),
data = dd_trainset)You will notice that I provided control parameters such as minsplit, minbucket, and cp to the rpart function. These parameters help us decide how the tree is constructed. You should try different settings to test whether you obtain different results.
Now, the easy part. Let’s see what the tree looks like.
dd.rp
#> n= 24195
#>
#> node), split, n, loss, yval, (yprob)
#> * denotes terminal node
#>
#> 1) root 24195 9200 Y (0.3801 0.6199)
#> 2) DEGREE_LEVEL=Non-Alumni 7856 212 N (0.9730 0.0270) *
#> 3) DEGREE_LEVEL=Graduate,Undergrad 16339 1550 Y (0.0950 0.9050)
#> 6) AGE< 25.5 1568 590 N (0.6237 0.3763)
#> 12) AGE>=22.5 1263 313 N (0.7522 0.2478)
#> 24) PREF_ADDRESS_TYPE=BUSN,HOME,OTR,Unknown 1191 256 N (0.7851 0.2149) *
#> 25) PREF_ADDRESS_TYPE=CAMP 72 15 Y (0.2083 0.7917) *
#> 13) AGE< 22.5 305 28 Y (0.0918 0.9082) *
#> 7) AGE>=25.5 14771 575 Y (0.0389 0.9611) *Let’s look at the complexity of the tree.
printcp(dd.rp)
#>
#> Classification tree:
#> rpart(formula = DONOR_IND ~ ., data = dd_trainset, method = "class",
#> control = rpart.control(minsplit = 2, minbucket = 2, cp = 0.001))
#>
#> Variables actually used in tree construction:
#> [1] AGE DEGREE_LEVEL
#> [3] PREF_ADDRESS_TYPE
#>
#> Root node error: 9197/24195 = 0.4
#>
#> n= 24195
#>
#> CP nsplit rel error xerror xstd
#> 1 0.808 0 1.0 1.0 0.008
#> 2 0.042 1 0.2 0.2 0.004
#> 3 0.027 2 0.1 0.1 0.004
#> 4 0.005 3 0.1 0.1 0.004
#> 5 0.001 4 0.1 0.1 0.004We can also visualize the complexity and model performance using the plotcp function.
plotcp(dd.rp)
Another way to see the model performance is to simply use the summary function.
summary(dd.rp)
#> Call:
#> rpart(formula = DONOR_IND ~ ., data = dd_trainset, method = "class",
#> control = rpart.control(minsplit = 2, minbucket = 2, cp = 0.001))
#> n= 24195
#>
#> CP nsplit rel error xerror xstd
#> 1 0.80809 0 1.000 1.000 0.00821
#> 2 0.04219 1 0.192 0.192 0.00440
#> 3 0.02707 2 0.150 0.150 0.00392
#> 4 0.00457 3 0.123 0.123 0.00357
#> 5 0.00100 4 0.118 0.120 0.00353
#>
#> Variable importance
#> DEGREE_LEVEL ALUMNUS_IND AGE
#> 39 39 22
#>
#> Node number 1: 24195 observations, complexity param=0.808
#> predicted class=Y expected loss=0.38 P(node) =1
#> class counts: 9197 14998
#> probabilities: 0.380 0.620
#> left son=2 (7856 obs) right son=3 (16339 obs)
#> Primary splits:
#> DEGREE_LEVEL splits as RLR, improve=8180.0, (0 missing)
#> ALUMNUS_IND splits as LR, improve=8090.0, (0 missing)
#> AGE < 25.5 to the left, improve=4910.0, (0 missing)
#> PARENT_IND splits as LR, improve= 182.0, (0 missing)
#> HAS_INVOLVEMENT_IND splits as LR, improve= 22.5, (0 missing)
#> Surrogate splits:
#> ALUMNUS_IND splits as LR, agree=0.998, adj=0.993, (0 split)
#> AGE < 25.5 to the left, agree=0.813, adj=0.424, (0 split)
#>
#> Node number 2: 7856 observations
#> predicted class=N expected loss=0.027 P(node) =0.325
#> class counts: 7644 212
#> probabilities: 0.973 0.027
#>
#> Node number 3: 16339 observations, complexity param=0.0422
#> predicted class=Y expected loss=0.095 P(node) =0.675
#> class counts: 1553 14786
#> probabilities: 0.095 0.905
#> left son=6 (1568 obs) right son=7 (14771 obs)
#> Primary splits:
#> AGE < 25.5 to the left, improve=970.000, (0 missing)
#> DEGREE_LEVEL splits as R-L, improve= 33.400, (0 missing)
#> PARENT_IND splits as LR, improve= 19.600, (0 missing)
#> HAS_INVOLVEMENT_IND splits as LR, improve= 2.790, (0 missing)
#> MARITAL_STATUS splits as RRRRLLR, improve= 0.846, (0 missing)
#>
#> Node number 6: 1568 observations, complexity param=0.0271
#> predicted class=N expected loss=0.376 P(node) =0.0648
#> class counts: 978 590
#> probabilities: 0.624 0.376
#> left son=12 (1263 obs) right son=13 (305 obs)
#> Primary splits:
#> AGE < 22.5 to the right, improve=214.0, (0 missing)
#> MARITAL_STATUS splits as LL--LRL, improve= 86.5, (0 missing)
#> PREF_ADDRESS_TYPE splits as LRLLL, improve= 39.5, (0 missing)
#> EMAIL_PRESENT_IND splits as LR, improve= 15.4, (0 missing)
#> DEGREE_LEVEL splits as R-L, improve= 10.2, (0 missing)
#>
#> Node number 7: 14771 observations
#> predicted class=Y expected loss=0.0389 P(node) =0.61
#> class counts: 575 14196
#> probabilities: 0.039 0.961
#>
#> Node number 12: 1263 observations, complexity param=0.00457
#> predicted class=N expected loss=0.248 P(node) =0.0522
#> class counts: 950 313
#> probabilities: 0.752 0.248
#> left son=24 (1191 obs) right son=25 (72 obs)
#> Primary splits:
#> PREF_ADDRESS_TYPE splits as LRLLL, improve=45.20, (0 missing)
#> MARITAL_STATUS splits as LL--LRL, improve=41.20, (0 missing)
#> EMAIL_PRESENT_IND splits as LR, improve=19.70, (0 missing)
#> DEGREE_LEVEL splits as R-L, improve=13.70, (0 missing)
#> HAS_INVOLVEMENT_IND splits as LR, improve= 6.47, (0 missing)
#>
#> Node number 13: 305 observations
#> predicted class=Y expected loss=0.0918 P(node) =0.0126
#> class counts: 28 277
#> probabilities: 0.092 0.908
#>
#> Node number 24: 1191 observations
#> predicted class=N expected loss=0.215 P(node) =0.0492
#> class counts: 935 256
#> probabilities: 0.785 0.215
#>
#> Node number 25: 72 observations
#> predicted class=Y expected loss=0.208 P(node) =0.00298
#> class counts: 15 57
#> probabilities: 0.208 0.792Using the rpart.plot function, we can visualize the tree.
rpart.plot(dd.rp, extra = 2, tweak = 1,
main = "Donor decision tree using the rpart package")
Let’s see how the model performed on the test data set. First, let’s store all the predictions.
predictions <- predict(dd.rp, dd_testset,
type = "class")Then, let’s create a cross-table of actual donors and non-donors and the predictions.
table(dd_testset$DONOR_IND, predictions)
#> predictions
#> N Y
#> N 3635 243
#> Y 199 6236Finally, let’s create a confusion matrix using the caret library.
confusionMatrix(
table(predictions, dd_testset$DONOR_IND),
positive = "Y")
#> Confusion Matrix and Statistics
#>
#>
#> predictions N Y
#> N 3635 199
#> Y 243 6236
#>
#> Accuracy : 0.957
#> 95% CI : (0.953, 0.961)
#> No Information Rate : 0.624
#> P-Value [Acc > NIR] : <2e-16
#>
#> Kappa : 0.908
#> Mcnemar's Test P-Value : 0.0408
#>
#> Sensitivity : 0.969
#> Specificity : 0.937
#> Pos Pred Value : 0.962
#> Neg Pred Value : 0.948
#> Prevalence : 0.624
#> Detection Rate : 0.605
#> Detection Prevalence : 0.628
#> Balanced Accuracy : 0.953
#>
#> 'Positive' Class : Y
#> The decision tree created using the example donor data observes 95.71% accuracy with primary node splits using features such as educational level (DEGREE_LEVEL), alumni type (ALUMNUS_IND), AGE, parent (PARENT_IND), and involvement indicator (HAS_INVOLVEMENT_IND).
Decision trees are widely used for building classification models based on their training flexibility, output readability, and ease of interpretation. The disadvantage of using decision trees is that they tend to be biased and over-fitted to noise (error) in your training data. To deal with this you can explore tree pruning, as well as other machine learning algorithms such as conditional inference trees, random forests, etc.
12.2.4.3 Conditional Inference Tree
The party package provides non-parametric classification (categorical) and regression (continuous) tree-based models. The specific type of tree created will depend on the outcome variable type (for example, nominal, ordinal, or numeric.), and the tree growth will be determined by statistical metrics.
Use the following command to install the party package (Hothorn et al. 2017):
# Install party
install.packages("party",
repos = 'http://cran.us.r-project.org')Let’s explore how to build a conditional inference tree using the party and caret library. Let’s load the libraries and prepare the data.
# Load party
library(party)
# Load caret library
library(caret)
# Load Data
donor_data <- read_csv("data/DonorSampleDataML.csv")
pred_vars <- c('MARITAL_STATUS', 'GENDER',
'ALUMNUS_IND', 'PARENT_IND',
'HAS_INVOLVEMENT_IND', 'DEGREE_LEVEL',
'PREF_ADDRESS_TYPE', 'EMAIL_PRESENT_IND',
'DONOR_IND')
donor_data <- select(donor_data,
pred_vars,
AGE)
# Set Seed for Repeatable Results
set.seed(777)
# Convert features to factor
donor_data <- mutate_at(donor_data,
.vars = c('MARITAL_STATUS', 'GENDER',
'ALUMNUS_IND', 'PARENT_IND',
'HAS_INVOLVEMENT_IND', 'DEGREE_LEVEL',
'PREF_ADDRESS_TYPE', 'EMAIL_PRESENT_IND',
'DONOR_IND'),
.funs = as.factor)
# Split 70% of donor_data into training
# data and 30% into test data
dd_index <- sample(2,
nrow(donor_data),
replace = TRUE,
prob = c(0.7, 0.3))
dd_trainset <- donor_data[dd_index == 1, ]
dd_testset <- donor_data[dd_index == 2, ]
# Confirm size of training and test datasets
dim(dd_trainset)
dim(dd_testset)Now, let’s use the ctree function to build a conditional inference tree.
dd.cit <- ctree(DONOR_IND ~ .,
data = dd_trainset)What does the conditional inference tree look like?
dd.citLet’s also see the summary of the mode.
summary(dd.cit)Next, let’s plot the conditional inference tree.
plot(dd.cit, main = "Donor Conditional Inference Tree")Now, let’s store the predictions and create a confusion matrix.
predictions <- predict(dd.cit, dd_testset)
# Generate cross-table for conditional inference tree
table(dd_testset$DONOR_IND, predictions)
# Create confusion matrix
confusionMatrix(
table(predictions, dd_testset$DONOR_IND),
positive = "Y")The ctree algorithm built a model with 95.71% accuracy with our test data.
12.2.4.4 C50
The C50 package is another popular tool for building decision trees (Kuhn and Quinlan 2017).
Let’s explore how to use the C50 algorithm to build a decision tree, but first, we should install the library.
# Install C50 package
install.packages("C50",
repos = 'http://cran.us.r-project.org')Next, let’s load and prep the data.
# Load readr
library(readr)
# Load dplyr
library(dplyr)
# Load lubridate
library(lubridate)
# Load C50 package
library(C50)
# Load caret library
library(caret)
# Set Seed for Repeatable Results
set.seed(123)
# Load Data
donor_data <- read_csv("data/DonorSampleDataML.csv")
# Drop 'ID', 'MEMBERSHIP_ID', etc.
pred_vars <- c('MARITAL_STATUS', 'GENDER',
'ALUMNUS_IND', 'PARENT_IND',
'HAS_INVOLVEMENT_IND', 'DEGREE_LEVEL',
'PREF_ADDRESS_TYPE', 'EMAIL_PRESENT_IND')
donor_data <- select(donor_data,
pred_vars,
DONOR_IND)
# Convert from character to numeric data type
convert_fac2num <- function(x){
as.numeric(as.factor(x))
}
# Convert features from factor to numeric
donor_data <- mutate_at(donor_data,
.vars = pred_vars,
.funs = convert_fac2num)
# Convert feature to factor
donor_data$DONOR_IND <- as.factor(donor_data$DONOR_IND)
# Split 70% of donor_data into training
# data and 30% into test data
dd_index <- sample(2,
nrow(donor_data),
replace = TRUE,
prob = c(0.7, 0.3))
dd_trainset <- donor_data[dd_index == 1, ]
dd_testset <- donor_data[dd_index == 2, ]
# Confirm size of training and test
# datasets
dim(dd_trainset)
#> [1] 24287 9
dim(dd_testset)
#> [1] 10221 9
# Create prop table
prop.table(table(dd_trainset$DONOR_IND))
#>
#> N Y
#> 0.377 0.623
prop.table(table(dd_testset$DONOR_IND))
#>
#> N Y
#> 0.383 0.617Using the prop.table function reveals there are about 62% donors and 38% non-donors in both the training and test datasets.
Now, let’s build the model using the C50 algorithm.
# Use C50 to build a decision tree model
donor_model <- C5.0(x = select(dd_trainset, -DONOR_IND),
y = dd_trainset$DONOR_IND)What does the tree look like?
donor_model
#>
#> Call:
#> C5.0.default(x = select(dd_trainset,
#> -DONOR_IND), y = dd_trainset$DONOR_IND)
#>
#> Classification Tree
#> Number of samples: 24287
#> Number of predictors: 8
#>
#> Tree size: 3
#>
#> Non-standard options: attempt to group attributesLet’s see the summary of the model.
summary(donor_model)
#>
#> Call:
#> C5.0.default(x = select(dd_trainset,
#> -DONOR_IND), y = dd_trainset$DONOR_IND)
#>
#>
#> C5.0 [Release 2.07 GPL Edition] Tue Jan 30 19:37:52 2018
#> -------------------------------
#>
#> Class specified by attribute `outcome'
#>
#> Read 24287 cases (9 attributes) from undefined.data
#>
#> Decision tree:
#>
#> DEGREE_LEVEL <= 1: Y (1710)
#> DEGREE_LEVEL > 1:
#> :...DEGREE_LEVEL <= 2: N (7840/206)
#> DEGREE_LEVEL > 2: Y (14737/1523)
#>
#>
#> Evaluation on training data (24287 cases):
#>
#> Decision Tree
#> ----------------
#> Size Errors
#>
#> 3 1729( 7.1%) <<
#>
#>
#> (a) (b) <-classified as
#> ---- ----
#> 7634 1523 (a): class N
#> 206 14924 (b): class Y
#>
#>
#> Attribute usage:
#>
#> 100.00% DEGREE_LEVEL
#>
#>
#> Time: 0.0 secsNow, let’s store the predictions and create a confusion matrix.
donor_predictions <- predict(donor_model,
dd_testset)
# Create confusion matrix
confusionMatrix(table(donor_predictions,
dd_testset$DONOR_IND),
positive = "Y")
#> Confusion Matrix and Statistics
#>
#>
#> donor_predictions N Y
#> N 3242 91
#> Y 676 6212
#>
#> Accuracy : 0.925
#> 95% CI : (0.92, 0.93)
#> No Information Rate : 0.617
#> P-Value [Acc > NIR] : <2e-16
#>
#> Kappa : 0.837
#> Mcnemar's Test P-Value : <2e-16
#>
#> Sensitivity : 0.986
#> Specificity : 0.827
#> Pos Pred Value : 0.902
#> Neg Pred Value : 0.973
#> Prevalence : 0.617
#> Detection Rate : 0.608
#> Detection Prevalence : 0.674
#> Balanced Accuracy : 0.907
#>
#> 'Positive' Class : Y
#> The C50 algorithm built a model with 92.50% accuracy with our test data. Let’s explore if we can improve the performance of the decision tree model using ensemble methods.
12.2.4.5 Ensemble Learning
Ensemble learning is a machine learning method that uses a combination of learning algorithms to improve predictive results. Boosting is a popular ensemble learning technique that reduces model bias by adding models that learn from the misclassification errors generated by other models.
Let’s explore how to use the boosting parameters within the C5.0 library to increase the performance of your decision tree models by adding multiple trials (models) with adaptive boosting.
12.2.4.6 Adaptive Boosting
We can create ensemble models by simply adding the number of boosting iterations to the trials parameter of the C50 function. Note that this code may take a long time to run.
# Set Seed for Repeatable Results
set.seed(123)
# Use C5.0 to build a decision tree model
# with n=20 trials of adaptive boosting
donor_model_boost20 <- C5.0(
x = select(dd_trainset, -DONOR_IND),
y = dd_trainset$DONOR_IND,
trials = 20)After the model is trained, we can see how well it performed by printing the model details and summary and looking at the confusion matrix.
# Show C5.0 decision tree with adaptive boosting
donor_model_boost20
# Examine C5.0 decision tree model with
# adaptive boosting
summary(donor_model_boost20)
# Apply decision tree to test data
donor_predictions_boost20 <- predict(donor_model_boost20,
dd_testset)
# Create confusion matrix
confusionMatrix(
table(donor_predictions_boost20, dd_testset$DONOR_IND),
positive = "Y)With our test data, the addition of 20 trials of adaptive boosting did not significantly increase the performance of the decision tree beyond 92.5%.
12.2.4.7 Random Forests
Random forests (also known as random decision forests) is an ensemble learning method that builds a collection of decision tree models and uses its output to correct any observed overfitting encountered during the training process.
The following example outlines how to use the randomForest library to build a random forest model to predict donor classification (Liaw, Wiener, and others 2002; Breiman et al. 2015).
# Install randomForest
install.packages("randomForest",
repos = 'http://cran.us.r-project.org')Let’s load the library and prep the data.
# Load randomForest
library(randomForest)
# Load Data
donor_data <- read_csv("data/DonorSampleDataML.csv")
# Drop 'ID', 'MEMBERSHIP_ID', etc.
pred_vars <- c('MARITAL_STATUS', 'GENDER',
'ALUMNUS_IND', 'PARENT_IND',
'HAS_INVOLVEMENT_IND', 'DEGREE_LEVEL',
'PREF_ADDRESS_TYPE', 'EMAIL_PRESENT_IND')
donor_data <- select(donor_data,
pred_vars,
DONOR_IND)
# Set Seed for Repeatable Results
set.seed(777)
# Convert features to factor
donor_data <- mutate_at(donor_data,
.vars = c(pred_vars,
'DONOR_IND'),
.funs = as.factor)
# Split 70% of donor_data into training
# data and 30% into test data
dd_index <- sample(2, nrow(donor_data), replace = TRUE,
prob = c(0.7, 0.3))
dd_trainset <- donor_data[dd_index == 1, ]
dd_testset <- donor_data[dd_index == 2, ]
# Confirm size of training and test
# datasets
dim(dd_trainset)
dim(dd_testset)Now let’s use the randomForest function to build a model.
dd.rf <- randomForest(DONOR_IND ~ .,
data = dd_trainset)
# Show randomForest details
dd.rf
# Examine model summary
summary(dd.rf)
# Print results
print(dd.rf)
# Measure predictor importance
importance(dd.rf)12.2.4.8 xgboost
Extreme gradient boosting (xgboost) is another ensemble learning method for classification and regression tasks. The xgboost library builds upon gradient boosting, which is an ensemble machine learning method that generates predictions using a collection of decision tree learners. xgboost is a popular machine learning tool in data mining competitions due to its speed and ability to execute parallel computations on a single machine (T. Chen et al. 2017).
To learn more about gradient boosting, you can refer to this article.
xgboost library and build an ensemble learning model to predict whether a constituent is a donor or non-donor based on available training (input) data.
12.2.5 Rule-Based Classification
Rule-based classification, similar to decision tree classification, is a machine learning method you can use to identify actionable patterns in your data. Rule-based models (also known as separate-and-conquer rule learners) select test conditions that help identify useful relationships in your data.
12.2.5.1 OneR
As seen in Section 3.2, the OneR (also known as One Rule) algorithm can be used to build a simple, yet accurate, model that strikes a balance between accuracy and interpretability (Holte 1993). OneR generates one rule for each predictor in your training data and then selects the rule with the smallest total error as its “one rule”.
Let’s explore how to build a classification model using the OneR algorithm. We will use the R version of the Weka machine learning package called RWeka (Witten and Frank 2005; Hornik, Buchta, and Zeileis 2009).
install.packages("RWeka",
repos = 'http://cran.us.r-project.org')# Load RWeka
library(RWeka)
# Set Seed for Repeatable Results
set.seed(777)Let’s build the OneR model using the OneR function from the RWeka library.
donor_OneR <- OneR(DONOR_IND ~ AGE + MARITAL_STATUS +
GENDER + ALUMNUS_IND + PARENT_IND +
HAS_INVOLVEMENT_IND + PREF_ADDRESS_TYPE +
EMAIL_PRESENT_IND,
data = donor_data)Let’s see what the model looks like.
donor_OneRWe can also see the summary of model performance using the summary function.
summary(donor_OneR)The OneR model observed an accuracy of 92.5%.
Let’s try another rule learning method to see if we can increase accuracy.
12.2.5.2 JRip
The JRip library implements a propositional rule learner, “Repeated Incremental Pruning to Produce Error Reduction” (RIPPER), as proposed in the article Fast Effective Rule Induction (Cohen 1995).
# Load RWeka
library(RWeka)
# Set Seed for Repeatable Results
set.seed(777)Let’s build the JRip model.
donor_JRip <- JRip(DONOR_IND ~ AGE + MARITAL_STATUS +
GENDER + ALUMNUS_IND + PARENT_IND +
HAS_INVOLVEMENT_IND +
PREF_ADDRESS_TYPE + EMAIL_PRESENT_IND,
data = donor_data)Let’s see what the JRIP rules look like.
donor_JRipAnd, finally, let’s see the model evaluation.
summary(donor_JRip)The JRip model generated 10 rules which correctly classified 95.6% of the test instances.
Let’s explore some other advanced machine learning methods that are used in various real-world applications.
12.2.5.3 Neural Networks
A neural network is a powerful machine learning technique that uses a network of computational models to provide iterative feedback and make predictions using input data. Neural networks, which simulate the interconnected signaling processes and activities of the human brain, are composed of input layers (training data), hidden layers (backpropagation layers that optimize input variable weights to improve predictive power), and output layers (predicted outcomes based on iterative feedback from input and hidden layers) (Nandeshwar 2006). A typical representation of a neural network is shown in Figure 12.1. Due to the complexity of neural network (NN) algorithms, they are often referred to as black box methods.
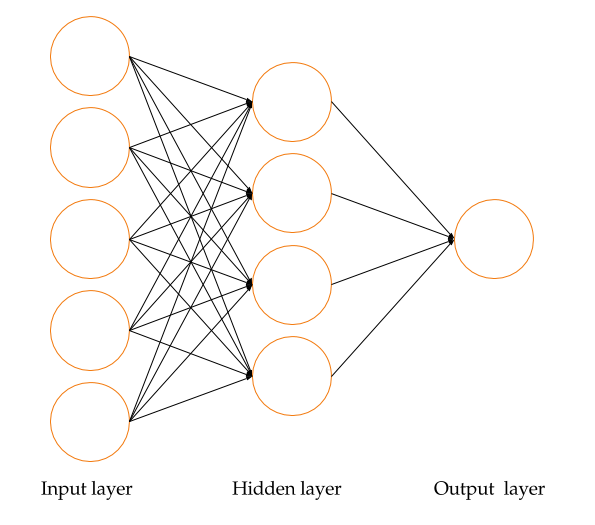
FIGURE 12.1: A neural network representation
Let’s explore how to build and implement an NN model using our synthetic donor data file. First, let’s load the libraries and prep the data.
# Load readr
library(readr)
# Load dplyr
library(dplyr)
# Load neuralnet package
library(neuralnet)
# Load caret package
library(caret)
# Set Seed for Repeatable Results
set.seed(123)
# Load Data
donor_data <- read_csv("data/DonorSampleDataML.csv")
# Drop 'ID' variable
pred_vars <- c('MARITAL_STATUS', 'GENDER',
'ALUMNUS_IND', 'PARENT_IND',
'HAS_INVOLVEMENT_IND', 'DEGREE_LEVEL',
'PREF_ADDRESS_TYPE', 'EMAIL_PRESENT_IND')
donor_data <- select(donor_data,
pred_vars,
DONOR_IND)
# Convert from character to numeric data type
convert_fac2num <- function(x){
as.numeric(as.factor(x))
}
# Convert features from factor to numeric
donor_data <- mutate_at(donor_data,
.vars = pred_vars),
.funs = convert_fac2num)
# Convert feature to factor
donor_data$DONOR_IND <- as.factor(donor_data$DONOR_IND)Then let’s divide 70% of the donor_data file into training data and 30% into test data.
dd_index <- sample(2,
nrow(donor_data),
replace = TRUE,
prob = c(0.7, 0.3))
dd_trainset <- donor_data[dd_index == 1, ]
dd_testset <- donor_data[dd_index == 2, ]
# Confirm size of training and test
# datasets
dim(dd_trainset)
dim(dd_testset)
# Select data variable predictors
dd_trainset <- select(dd_trainset[1:5000,],
pred_vars)
dd_testset <- select(dd_testset[1:5000,],
pred_vars)
# Define min-max normalization function
min_max <- function(x) {
return((x - min(x))/(max(x) - min(x)))
}
# Apply min-max normalization to training
# and test datasets except DONOR_IND
dd_trainset_n <- mutate_at(dd_trainset,
.vars = pred_vars,
.funs = min_max)
dd_testset_n <- mutate_at(dd_testset,
.vars = pred_vars,
.funs = min_max)
# Recombine and prepare datasets for Neural Network (NN)
dd_trainset_n <- cbind(dd_trainset_n, dd_trainset[9])
dd_trainset_n <- cbind(dd_trainset_n,
dd_trainset_n$DONOR_IND == 'Y')
dd_trainset_n <- cbind(dd_trainset_n,
dd_trainset_n$DONOR_IND == 'N')
names(dd_trainset_n)[10] <- 'donor'
names(dd_trainset_n)[11] <- 'non_donor'
# Remove DONOR_IND for NN
dd_trainset_n <- dd_trainset_n[-c(9)]
# Prepare testset for NN
dd_testset_n <- cbind(dd_testset_n,dd_testset[9])
# Build NN model
dd_pred_nn <- neuralnet(donor + non_donor ~ AGE + MARITAL_STATUS
+ GENDER + ALUMNUS_IND + PARENT_IND + HAS_INVOLVEMENT_IND +
DEGREE_LEVEL + EMAIL_PRESENT_IND,
data = dd_trainset_n, hidden = 1, threshold = 0.01)
# Examine NN model
dd_pred_nn$result.matrix
# Plot NN model
plot(dd_pred_nn)
# Make predictions using NN model
donor_prediction <- compute(dd_pred_nn, dd_testset_n[-9])$net.result
# Measure correlation predicted giving and true value
cor(donor_prediction[,1], as.numeric(dd_testset_n$DONOR_IND))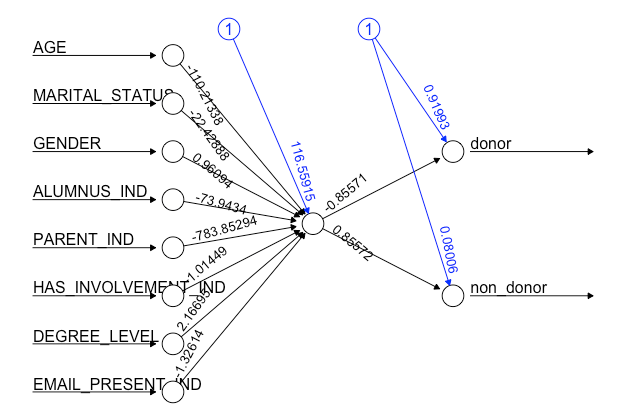
FIGURE 12.2: Donor neural network using neuralnet
In this example, we covered how to build an NN model, which performed relatively well with an 82% correlation between the predicted classification (donor versus non-donor) outcomes and true values relative to our test donor data.
Overall, NN models perform well with learning non-linear functions and handling multiple output variables at the same time. As you might have already figured out, the primary disadvantages of NNs are that they (1) require additional training time and (2) can be more difficult to interpret than some of the other traditional and less complex machine learning algorithms previously covered.
Let’s take a look at another powerful machine learning method you will likely encounter on your data science journey.
12.2.5.4 Support Vector Machines
Support vector machines (SVM) (also known as support vector networks) is another powerful black box algorithm commonly used with supervised learning tasks such as classification and regression.
SVM uses kernel functions to map input data into high-dimensional feature (variable) spaces to efficiently perform non-linear classification. Machine learning algorithms capable of operating with kernel functions and statistical learning parameters include SVM, principal components analysis (PCA), ridge regression, spectral clustering, linear adaptive filters, and many others.
While the inner workings of NN and SVM algorithms are beyond the scope of this book, we will provide an SVM recipe you can use and test while exploring other machine learning methods and applications with your data.
# Load readr
library(readr)
# Load dplyr
library(dplyr)
# Load e1071
library(e1071)
# Load caret package
library(caret)
# Set Seed for Repeatable Results
set.seed(123)
# Load Data
donor_data <- read_csv("data/DonorSampleDataML.csv")
# Drop 'ID', 'MEMBERSHIP_ID', etc.
pred_vars <- c('MARITAL_STATUS', 'GENDER',
'ALUMNUS_IND', 'PARENT_IND',
'HAS_INVOLVEMENT_IND', 'DEGREE_LEVEL',
'PREF_ADDRESS_TYPE', 'EMAIL_PRESENT_IND',
'DONOR_IND')
donor_data <- select(donor_data,
pred_vars,
AGE)
# Convert features to factor
donor_data <- mutate_at(donor_data,
.vars = pred_vars,
.funs = as.factor)
# Split 70% of donor_data into training
# data and 30% into test data
dd_index <- sample(2, nrow(donor_data), replace = TRUE,
prob = c(0.7, 0.3))
dd_trainset <- donor_data[dd_index == 1, ]
dd_testset <- donor_data[dd_index == 2, ]
# Confirm size of training and test
# datasets
dim(dd_trainset)
dim(dd_testset)Finally, let’s build the model.
dd_svm <- svm(DONOR_IND ~ MARITAL_STATUS + GENDER +
ALUMNUS_IND + PARENT_IND +
HAS_INVOLVEMENT_IND + DEGREE_LEVEL +
EMAIL_PRESENT_IND + AGE,
data = dd_trainset,
kernel = "radial",
cost = 1,
gamma = 1/ncol(dd_trainset))Let’s see what the model looks like.
dd_svmNext, let’s store the predictions and create a confusion matrix.
donor_prediction <- predict(dd_svm,
select(dd_testset,
pred_vars))
# Create SVM crosstab
svm.crosstab <- table(donor_prediction, dd_testset$DONOR_IND)
# Confusion Matrix
confusionMatrix(svm.crosstab, positive = "Y")In this example, we built an SVM model using the e1071 package and our donor file. The SVM model predicted donor versus non-donor classification with an accuracy of 94%, which suggests it is a useful machine learning algorithm to keep in mind for future predictive modeling projects.
SVMs, similar to neural networks, can efficiently handle non-linear classification problems, which makes it useful with text categorization, image segmentation, and other science-based applications. You can refer to this article if you would like to learn more.
Now, let’s turn our attention to another exciting topic in machine learning concepts: deep learning.
12.2.5.5 Deep Learning with TensorFlow
The field of data analytics is dynamic, so it is important to recognize that the machine learning algorithms we are using today will continue to improve and evolve in terms of efficiency and capability.
Deep learning (also known as deep structured learning or hierarchical learning) is a type of machine learning focused on learning data representations and feature learning rather than individual or specific tasks. Feature learning (also known as representation learning) can be supervised, semi-supervised, or unsupervised.
Deep learning architectures include deep neural networks, deep belief networks, and recurrent neural networks. Real-world applications using deep learning include computer vision, speech recognition, machine translation, natural language processing, image recognition, and so on.
The following recipe introduces how to implement a deep neural network using TensorFlow, which is an open source software library (originally developed at Google) for complex computation by constructing network graphs of mathematical operations and data (Abadi et al. 2016; Cheng et al. 2017). Tang et al. (2017) developed an R interface to the TensorFlow API for our use.
Before we use this library, we need to install it. Since this is a very recent library, we will install it from GitHub directly.
devtools::install_github("rstudio/tfestimators")
library(tfestimators)Although we’ve installed the library, we don’t have the actual compiled code for TensorFlow, which we need to install using the install_tensorlfow() command that came with the tfestimators package.
install_tensorflow()When you try to run it, you may face an error like the following:
#> Error: Prerequisites for installing
#> TensorFlow not available. Execute the
#> following at a terminal to install the
#> prerequisites: $ sudo
#> /usr/local/bin/pip install --upgrade
#> virtualenvI was able to fix the error by running the above command on a Mac or Linux machine. On Windows, you may need further troubleshooting. After installing the prerequisites, you can try installing TensorFlow again.
install_tensorflow()Next, let’s load the data.
library(readr)
library(dplyr)
donor_data <- read_csv("data/DonorSampleDataCleaned.csv")The TensorFlow library doesn’t tolerate missing values; therefore, we will replace missing factor values with modes and missing numeric values with medians.
# function copied from
# https://stackoverflow.com/a/8189441/934898
my_mode <- function(x) {
ux <- unique(x)
ux[which.max(tabulate(match(x, ux)))]
}
donor_data <- donor_data %>%
mutate_if(is.numeric,
.funs = funs(
ifelse(is.na(.),
median(., na.rm = TRUE),
.))) %>%
mutate_if(is.character,
.funs = funs(
ifelse(is.na(.),
my_mode(.),
.)))Next, we need to convert the character variables to factors.
predictor_cols <- c("MARITAL_STATUS", "GENDER",
"ALUMNUS_IND", "PARENT_IND",
"WEALTH_RATING", "PREF_ADDRESS_TYPE")
# Convert feature to factor
donor_data <- mutate_at(donor_data,
.vars = predictor_cols,
.funs = as.factor)Now, we need to let TensorFlow know about the column types. For factor columns, we need to specify all the values contained in those columns using the column_categorical_with_vocabulary_list function. Then, using the column_indicator function, we convert each of the factor values in a column to its own column with 0s and 1s — this process is known as one hot encoding. For example, for the GENDER column say, we have two possible values of male and female. The one hot encoding process will create two columns, one for male and the other for female. Each of these columns will contain either 0 or 1, depending on the data value the GENDER column contained.
feature_cols <- feature_columns(
column_indicator(
column_categorical_with_vocabulary_list(
"MARITAL_STATUS",
vocabulary_list = unique(donor_data$MARITAL_STATUS))),
column_indicator(
column_categorical_with_vocabulary_list(
"GENDER",
vocabulary_list = unique(donor_data$GENDER))),
column_indicator(
column_categorical_with_vocabulary_list(
"ALUMNUS_IND",
vocabulary_list = unique(donor_data$ALUMNUS_IND))),
column_indicator(
column_categorical_with_vocabulary_list(
"PARENT_IND",
vocabulary_list = unique(donor_data$PARENT_IND))),
column_indicator(
column_categorical_with_vocabulary_list(
"WEALTH_RATING",
vocabulary_list = unique(donor_data$WEALTH_RATING))),
column_indicator(
column_categorical_with_vocabulary_list(
"PREF_ADDRESS_TYPE",
vocabulary_list = unique(donor_data$PREF_ADDRESS_TYPE))),
column_numeric("AGE"))After we’ve created the column types, let’s partition the data set into train and test datasets.
row_indices <- sample(1:nrow(donor_data),
size = 0.8 * nrow(donor_data))
donor_data_train <- donor_data[row_indices, ]
donor_data_test <- donor_data[-row_indices, ]The TensorFlow package then requires that we create an input function with the listing of input and output variables.
donor_pred_fn <- function(data) {
input_fn(data,
features = c("AGE", "MARITAL_STATUS",
"GENDER", "ALUMNUS_IND",
"PARENT_IND", "WEALTH_RATING",
"PREF_ADDRESS_TYPE"),
response = "DONOR_IND")
}Finally, we can use the prepared data set as well as the input function to build a deep-learning classifier. We will create three hidden layers with 80, 40, and 30 nodes, respectively.
classifier <- dnn_classifier(
feature_columns = feature_cols,
hidden_units = c(80, 40, 30),
n_classes = 2,
label_vocabulary = c("N", "Y"))Using the train function we will build the classifier.
train(classifier,
input_fn = donor_pred_fn(donor_data_train))We will next predict the values using the model for the test data set as well as the full data set.
predictions_test <- predict(
classifier,
input_fn = donor_pred_fn(donor_data_test))
predictions_all <- predict(
classifier,
input_fn = donor_pred_fn(donor_data))Similarly, we will evaluate the model for both the test data and the full data set. You can see the evaluation using the test data in Table 12.1 and using the full data sets in Table 12.2.
evaluation_test <- evaluate(
classifier,
input_fn = donor_pred_fn(donor_data_test))
evaluation_all <- evaluate(
classifier,
input_fn = donor_pred_fn(donor_data))| Measure | Value |
|---|---|
| accuracy | 84.34 |
| accuracy_baseline | 0.63 |
| auc | 216.00 |
| auc_precision_recall | 0.51 |
| average_loss | 0.62 |
| global_step | 0.63 |
| label/mean | 0.66 |
| loss | 0.63 |
| prediction/mean | 0.63 |
| Measure | Value |
|---|---|
| accuracy | 84.87 |
| accuracy_baseline | 0.62 |
| auc | 216.00 |
| auc_precision_recall | 0.51 |
| average_loss | 0.62 |
| global_step | 0.62 |
| label/mean | 0.66 |
| loss | 0.62 |
| prediction/mean | 0.62 |
12.2.6 Regression
Regression is a supervised machine learning method that analyzes and estimates the relationships among variables. Regression is widely used for prediction and forecasting tasks. Regression models are used to make numerical predictions of a continuous variable or can be used to make classification (categorical) predictions of a discrete response (dependent) variable.
12.2.6.1 Linear Regression
Linear regression is the simplest type of regression and is suitable if there is a linear relationship between the predictor (independent) and response (dependent) variables in your data. Otherwise, you should consider and explore other approaches (quadratic, polynomial, logistic, and so on).
Let’s explore how to fit a linear model using the example donor file. We will use the age variable to predict the total giving of a donor. Let’s load the data.
# Load readr
library(readr)
# Load dplyr
library(dplyr)
# Load Data
donor_data <- read_csv("data/DonorSampleDataML.csv")
# Set Seed for Repeatable Results
set.seed(777)Let’s look at the relationship between age and total giving by plotting that data.
with(donor_data, plot(AGE, TotalGiving))
To create the linear regression model, we will use the in-built lm function.
giving_model <- lm(TotalGiving ~ AGE,
data = donor_data)Next, to see the fit of the model, we will plot the regression line using the abline function.
with(donor_data, plot(AGE, TotalGiving))
abline(giving_model, col = "red")
Finally, we will see the model performance using the summary function.
summary(giving_model)
#>
#> Call:
#> lm(formula = TotalGiving ~ AGE, data = donor_data)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -31428 -3579 -3379 3382 12199436
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -13620.8 1944.5 -7.00 2.5e-12
#> AGE 409.5 47.3 8.66 < 2e-16
#>
#> (Intercept) ***
#> AGE ***
#> ---
#> Signif. codes:
#> 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 114000 on 34506 degrees of freedom
#> Multiple R-squared: 0.00217, Adjusted R-squared: 0.00214
#> F-statistic: 75 on 1 and 34506 DF, p-value: <2e-16The summary(giving_model) command produces various types of information you can use to evaluate the model’s performance in greater detail.
The residuals provide summary statistics for the predictions errors. For example, the inter-quartile range (IQR) of the prediction errors (residuals) in this model range between -3,578 over the true value and 3,382 under the true value.
The “***" next to the AGE coefficient indicates that the AGE feature (variable) is extremely unlikely to be unrelated to the “TotalGiving” response variable, which suggests we should keep this variable in mind for feature selection when building other models.
The R-squared value (also known as the coefficient of determination) describes the proportion of variance in TotalGiving (dependent variable) that is explained by the model. Broadly speaking, the closer that value is to 1.0, the better the model explains the observed variance in the input data. In this instance, the adjusted R-squared value is 0.002, which indicates that only 0.2% of the variance is explained by the model and suggests we should try a different approach. Most real-world applications using regression models include more than one independent variable for numeric prediction tasks, which we will further explore in the next recipe.
12.2.6.2 Multiple Linear Regression
Multiple linear regression models the relationships between two or more independent (explanatory) variables and a numerical dependent (response) variable by fitting a linear equation to observed data.
Let’s build on the previous example and explore how to use multiple independent variables to fit a multiple linear regression model to predict the current fiscal year’s giving.
# Load readr
library(readr)
# Load dplyr
library(dplyr)
# Load Data
donor_data <- read_csv("data/DonorSampleDataML.csv")
# Drop 'ID', 'MEMBERSHIP_ID', etc.
pred_vars <- c('MARITAL_STATUS', 'GENDER',
'ALUMNUS_IND', 'PARENT_IND',
'HAS_INVOLVEMENT_IND', 'DEGREE_LEVEL',
'PREF_ADDRESS_TYPE', 'EMAIL_PRESENT_IND')
donor_data <- select(donor_data,
pred_vars,
AGE,
PrevFYGiving,
PrevFY1Giving,
PrevFY2Giving,
PrevFY3Giving,
PrevFY4Giving,
CurrFYGiving)
# Set Seed for Repeatable Results
set.seed(777)
# Convert features to factor
donor_data <- mutate_at(donor_data,
.vars = pred_vars,
.funs = as.factor)Now that our data is loaded and prepared, let’s build the multiple linear regression model.
# Fit multiple linear regression model
giving_mlg_model <- lm(CurrFYGiving ~ .,
data = donor_data)Let’s see what the model performance looks like.
summary(giving_mlg_model)
#>
#> Call:
#> lm(formula = CurrFYGiving ~ ., data = donor_data)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -402697 -83 62 150 1147081
#>
#> Coefficients:
#> Estimate Std. Error
#> (Intercept) 2.42e+02 1.35e+03
#> MARITAL_STATUSMarried -6.95e+02 9.07e+02
#> MARITAL_STATUSNever Married -3.84e+02 4.80e+03
#> MARITAL_STATUSSeparated -2.06e+03 2.87e+03
#> MARITAL_STATUSSingle -8.57e+02 9.19e+02
#> MARITAL_STATUSUnknown -9.45e+02 9.08e+02
#> MARITAL_STATUSWidowed -9.16e+02 1.12e+03
#> GENDERMale 4.82e+00 9.04e+01
#> GENDERUnknown -4.09e+00 2.23e+02
#> ALUMNUS_INDY -2.00e+00 9.39e+02
#> PARENT_INDY -3.76e+02 2.15e+02
#> HAS_INVOLVEMENT_INDY 9.58e+01 1.08e+02
#> DEGREE_LEVELNon-Alumni 3.26e+02 9.53e+02
#> DEGREE_LEVELUndergrad 1.61e+02 1.81e+02
#> PREF_ADDRESS_TYPECAMP 1.87e+02 4.18e+02
#> PREF_ADDRESS_TYPEHOME 1.09e+01 2.67e+02
#> PREF_ADDRESS_TYPEOTR -1.21e+02 1.00e+03
#> PREF_ADDRESS_TYPEUnknown 8.57e+01 2.92e+02
#> EMAIL_PRESENT_INDY -1.98e+01 9.67e+01
#> AGE 9.47e+00 3.73e+00
#> PrevFYGiving -8.50e-02 8.83e-04
#> PrevFY1Giving 3.09e-03 4.21e-03
#> PrevFY2Giving 4.48e+00 1.46e-02
#> PrevFY3Giving 1.60e-03 2.55e-02
#> PrevFY4Giving -2.28e-02 6.56e-03
#> t value Pr(>|t|)
#> (Intercept) 0.18 0.8581
#> MARITAL_STATUSMarried -0.77 0.4432
#> MARITAL_STATUSNever Married -0.08 0.9362
#> MARITAL_STATUSSeparated -0.72 0.4728
#> MARITAL_STATUSSingle -0.93 0.3513
#> MARITAL_STATUSUnknown -1.04 0.2984
#> MARITAL_STATUSWidowed -0.82 0.4121
#> GENDERMale 0.05 0.9575
#> GENDERUnknown -0.02 0.9854
#> ALUMNUS_INDY 0.00 0.9983
#> PARENT_INDY -1.75 0.0802 .
#> HAS_INVOLVEMENT_INDY 0.89 0.3750
#> DEGREE_LEVELNon-Alumni 0.34 0.7321
#> DEGREE_LEVELUndergrad 0.89 0.3737
#> PREF_ADDRESS_TYPECAMP 0.45 0.6542
#> PREF_ADDRESS_TYPEHOME 0.04 0.9674
#> PREF_ADDRESS_TYPEOTR -0.12 0.9037
#> PREF_ADDRESS_TYPEUnknown 0.29 0.7695
#> EMAIL_PRESENT_INDY -0.21 0.8375
#> AGE 2.54 0.0111 *
#> PrevFYGiving -96.22 <2e-16 ***
#> PrevFY1Giving 0.73 0.4627
#> PrevFY2Giving 307.64 <2e-16 ***
#> PrevFY3Giving 0.06 0.9499
#> PrevFY4Giving -3.48 0.0005 ***
#> ---
#> Signif. codes:
#> 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 8160 on 34483 degrees of freedom
#> Multiple R-squared: 0.735, Adjusted R-squared: 0.735
#> F-statistic: 3.99e+03 on 24 and 34483 DF, p-value: <2e-16Let’s also plot the model.
par(mfrow = c(2, 2))
plot(giving_mlg_model)
#> Warning in sqrt(crit * p * (1 - hh)/hh): NaNs
#> produced
#> Warning in sqrt(crit * p * (1 - hh)/hh): NaNs
#> produced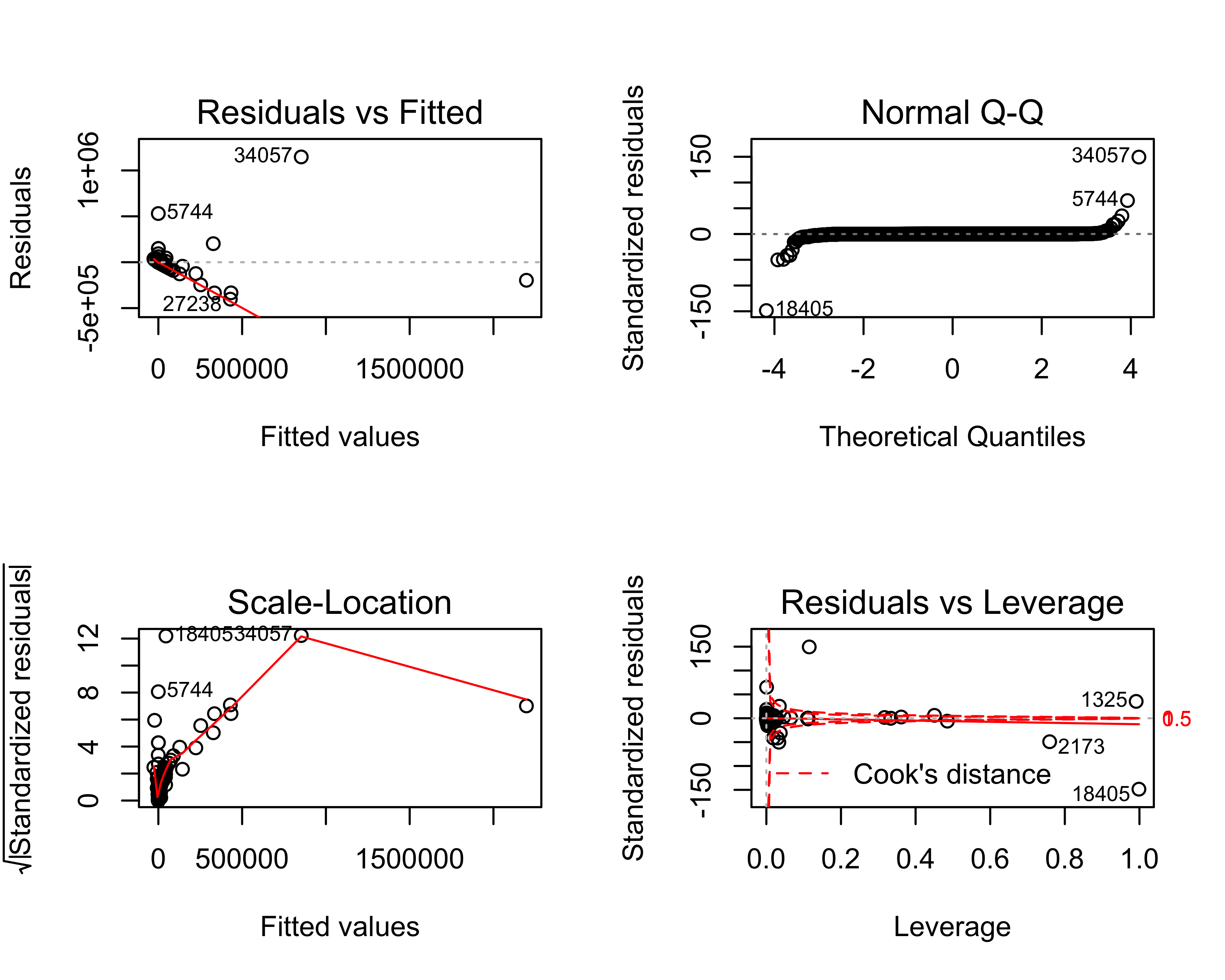
Based on the summary(giving_mlg_model) output, the multiple linear regression model does a much better job of predicting current fiscal year giving than our previous linear regression model, which predicted total giving. Specifically, the inter-quartile range (IQR) of the prediction errors (residuals) in this multiple linear regression model range between $83 over the true value and $150 under the true value. It makes sense that the Age and PrevFY*Giving variables are correlated with current FY’s giving.
The R-squared value (coefficient of determination) describes the proportion of variance in CurrFYGiving (dependent variable) that is explained by the model. As previously mentioned, the closer that value is to 1.0, the better the model explains the observed variance in the input data. In this instance, the adjusted R-squared value is 0.735, which indicates that approximately 74% of the observed variance in CurrFYGiving is explained by this model.
In the Predicting Gift Size and [Finding Prospects for Mail Solicitations] chapters, we will further explore how to use regression analysis to predict gift size and select prospects for mailing solicitations.
12.2.6.3 Logistic Regression
Logistic regression models the relationships between one or more independent (explanatory) variables and a dependent (outcome) variable that is categorical. Binary logistic regression is a common type of logistic regression where the dependent (response) variable is binary or dichotomous (that is, takes only one of two values such as giving/not giving to attended/not attended).
Let’s explore how to create a logistic regression model to predict whether a constituent will be a donor or non-donor based on input data. Let’s load and prep the data.
# Load readr
library(readr)
# Load dplyr
library(dplyr)
# Load caret package
library(caret)
# Load Data
donor_data <- read_csv("data/DonorSampleDataML.csv")
# Drop 'ID', 'MEMBERSHIP_ID', etc.
pred_vars <- c('MARITAL_STATUS', 'GENDER',
'ALUMNUS_IND', 'PARENT_IND',
'HAS_INVOLVEMENT_IND', 'DEGREE_LEVEL',
'PREF_ADDRESS_TYPE', 'EMAIL_PRESENT_IND',
'DONOR_IND')
donor_data <- select(donor_data,
pred_vars,
AGE)
# Set Seed for Repeatable Results
set.seed(777)
# Convert features to factor
donor_data <- mutate_at(donor_data,
.vars = pred_vars,
.funs = as.factor)Let’s split 70% of the data into training and the rest into test data.
dd_index <- sample(2,
nrow(donor_data),
replace = TRUE,
prob = c(0.7, 0.3))
dd_trainset <- donor_data[dd_index == 1, ]
dd_testset <- donor_data[dd_index == 2, ]
# Confirm size of training and test
# datasets
dim(dd_trainset)
#> [1] 24195 10
dim(dd_testset)
#> [1] 10313 10Now, let’s fit the logistic regression model.
giving_lr_model <- glm(DONOR_IND ~ .,
data = dd_trainset,
family = "binomial")Let’s see how the model performed.
summary(giving_lr_model)
#>
#> Call:
#> glm(formula = DONOR_IND ~ ., family = "binomial", data = dd_trainset)
#>
#> Deviance Residuals:
#> Min 1Q Median 3Q Max
#> -3.499 -0.093 0.144 0.419 3.774
#>
#> Coefficients:
#> Estimate Std. Error
#> (Intercept) 29.54367 759.25367
#> MARITAL_STATUSMarried -0.72166 0.72249
#> MARITAL_STATUSNever Married -1.06711 5.42223
#> MARITAL_STATUSSeparated 3.18970 2.45027
#> MARITAL_STATUSSingle -0.44973 0.72734
#> MARITAL_STATUSUnknown -0.35620 0.72183
#> MARITAL_STATUSWidowed -0.47427 0.81945
#> GENDERMale -0.12138 0.05532
#> GENDERUnknown -0.20367 0.13083
#> ALUMNUS_INDY -14.80159 743.91015
#> PARENT_INDY 2.34458 0.21162
#> HAS_INVOLVEMENT_INDY 0.20372 0.06838
#> DEGREE_LEVELNon-Alumni -37.01917 759.25330
#> DEGREE_LEVELUndergrad -16.04282 151.86597
#> PREF_ADDRESS_TYPECAMP 1.09975 0.26711
#> PREF_ADDRESS_TYPEHOME 0.04452 0.17423
#> PREF_ADDRESS_TYPEOTR 0.60209 0.79611
#> PREF_ADDRESS_TYPEUnknown -0.23205 0.18866
#> EMAIL_PRESENT_INDY 0.07867 0.05961
#> AGE 0.09819 0.00252
#> z value Pr(>|z|)
#> (Intercept) 0.04 0.9690
#> MARITAL_STATUSMarried -1.00 0.3179
#> MARITAL_STATUSNever Married -0.20 0.8440
#> MARITAL_STATUSSeparated 1.30 0.1930
#> MARITAL_STATUSSingle -0.62 0.5364
#> MARITAL_STATUSUnknown -0.49 0.6217
#> MARITAL_STATUSWidowed -0.58 0.5627
#> GENDERMale -2.19 0.0282 *
#> GENDERUnknown -1.56 0.1195
#> ALUMNUS_INDY -0.02 0.9841
#> PARENT_INDY 11.08 < 2e-16 ***
#> HAS_INVOLVEMENT_INDY 2.98 0.0029 **
#> DEGREE_LEVELNon-Alumni -0.05 0.9611
#> DEGREE_LEVELUndergrad -0.11 0.9159
#> PREF_ADDRESS_TYPECAMP 4.12 3.8e-05 ***
#> PREF_ADDRESS_TYPEHOME 0.26 0.7983
#> PREF_ADDRESS_TYPEOTR 0.76 0.4495
#> PREF_ADDRESS_TYPEUnknown -1.23 0.2187
#> EMAIL_PRESENT_INDY 1.32 0.1869
#> AGE 38.99 < 2e-16 ***
#> ---
#> Signif. codes:
#> 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 32136.9 on 24194 degrees of freedom
#> Residual deviance: 9839.7 on 24175 degrees of freedom
#> AIC: 9880
#>
#> Number of Fisher Scoring iterations: 17Next, let’s make predictions and store them.
predictions <- predict(giving_lr_model,
newdata = dd_testset,
type = "response")
preds <- as.factor(ifelse(predictions > 0.5,
"Y",
"N"))Finally, let’s see the confusion matrix.
confusionMatrix(table(preds, dd_testset$DONOR_IND),
positive = "Y")
#> Confusion Matrix and Statistics
#>
#>
#> preds N Y
#> N 3198 106
#> Y 680 6329
#>
#> Accuracy : 0.924
#> 95% CI : (0.918, 0.929)
#> No Information Rate : 0.624
#> P-Value [Acc > NIR] : <2e-16
#>
#> Kappa : 0.833
#> Mcnemar's Test P-Value : <2e-16
#>
#> Sensitivity : 0.984
#> Specificity : 0.825
#> Pos Pred Value : 0.903
#> Neg Pred Value : 0.968
#> Prevalence : 0.624
#> Detection Rate : 0.614
#> Detection Prevalence : 0.680
#> Balanced Accuracy : 0.904
#>
#> 'Positive' Class : Y
#> Logistic regression performs with an accuracy of 92.4%, which is on par with the performance of the KNN, C50, and OneR algorithms we previously explored.
Now that we have covered supervised learning algorithms, let’s explore another machine learning method that does not require labeled input (training) data.
12.3 Unsupervised Learning
Unsupervised learning is a type of machine learning that draws inferences and extracts hidden structure from unlabeled input data. In contrast to supervised learning, unsupervised learning does not require labeled data for training because there is no predicted outcome. Instead, unsupervised learning algorithms rely on the similarity (distance) metrics between data features (variables) to organize data into groupings known as clusters.
The de facto unsupervised learning method is cluster analysis, which can be used to find emergent patterns in your data. There are many different types of clustering methods, including hierarchical clustering, k-means clustering, model-based clustering, and density-based clustering.
K-means is one of the most popular and widely used clustering algorithms, which randomly assigns each observation (data point) to a cluster and calculates the centroid (that is, cluster center) of each cluster. The kmeans algorithm continues to reassign data points to the cluster with the closest centroid value and iteratively updates cluster centroid values until the cluster variation cannot be further minimized.
In the following recipe, we will explore how to use the kmeans algorithm to perform cluster analysis and reveal hidden groupings in the donor data file.
12.3.1 K-Means
Let’s load and prep the data first.
# Load readr
library(readr)
# Load dplyr
library(dplyr)
# Load ggplot2
library(ggplot2)
library(scales)
# Set Seed for Repeatable Results
set.seed(123)
# Load Data
donor_data <- read_csv("data/DonorSampleDataML.csv")
pred_vars <- c('MARITAL_STATUS', 'GENDER',
'ALUMNUS_IND', 'PARENT_IND',
'HAS_INVOLVEMENT_IND', 'DEGREE_LEVEL',
'PREF_ADDRESS_TYPE', 'EMAIL_PRESENT_IND')
# Convert from character to numeric data type
convert_fac2num <- function(x){
as.numeric(as.factor(x))
}
# Convert features from factor to numeric
donor_data <- mutate_at(donor_data,
.vars = pred_vars,
.funs = convert_fac2num)
# Convert feature to factor
donor_data$DONOR_IND <- as.factor(donor_data$DONOR_IND)Since we want to use the DONOR_IND variable for clustering, we will convert that variable into two columns with 1s and 0s (that is, one hot code that variable).
donor_data <- mutate(donor_data,
donor = ifelse(DONOR_IND =='Y', 1, 0),
non_donor = ifelse(DONOR_IND =='N', 1, 0))Let’s select the variables we want to use for cluster analysis.
donor_data <- select(donor_data,
pred_vars,
AGE,
TotalGiving,
donor,
non_donor)Now, let’s build the clusters using the kmeans algorithm and select five clusters.
dd_kmeans <- kmeans(x = donor_data,
centers = 5,
nstart = 10)We’ll look at the cluster distribution, but first let’s convert the cluster numbers to factors and save it to the donor_data data frame.
donor_data$cluster <- as.factor(dd_kmeans$cluster)
# Plot kmeans model
ggplot(donor_data, aes(x = AGE, y = TotalGiving, color = cluster)) +
geom_jitter() +
scale_y_log10(labels = dollar) +
theme_bw(base_size = 12) +
xlab("Donor Age") + ylab("Log (Total Giving)") +
ggtitle("Donor Cluster Analysis") +
scale_color_manual("Cluster Assignment",
labels = c("A", "B", "C", "D", "E"),
values = c("royalblue2", "orange",
"magenta3", "gray76", "limegreen")) +
theme(plot.title = element_text(hjust = 0.5))
#> Warning: Transformation introduced infinite values
#> in continuous y-axis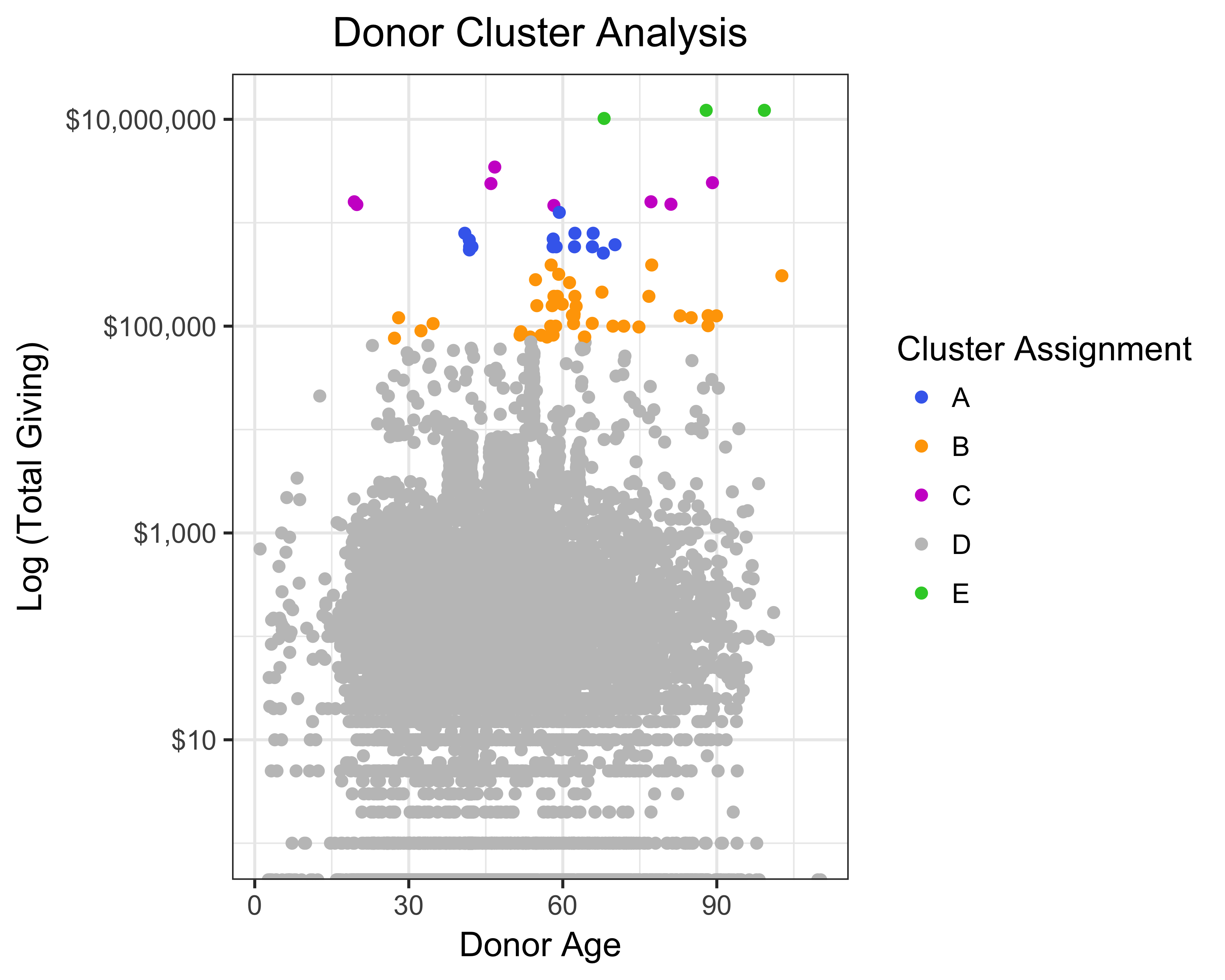
The kmeans algorithm requires that you specify k, which is the number of clusters used to organize the input data. In this example, the value of k was chosen arbitrarily, but you may also have domain expertise or business requirements informing your cluster number criteria. For smaller datasets, a rule of thumb is to define k as the square root of n/2 (that, sqrt(n/2)), where n equals the total number of observations in your dataset. For larger datasets, k can become very large and therefore difficult to manage and interpret with this cluster-selection approach. For additional details about choosing the optimal number of clusters, you can read the following article.
Once you’ve built your clustering model, it is useful to explore, compare, and describe your cluster segments using the average or median values for each of the clusters. For example, we built a cluster model with the cluster means shown in Figure 12.3 and added descriptions. This way, you can estimate the attributes of individual members of a cluster. You can further use these attributes or labels for marketing or segmentation.
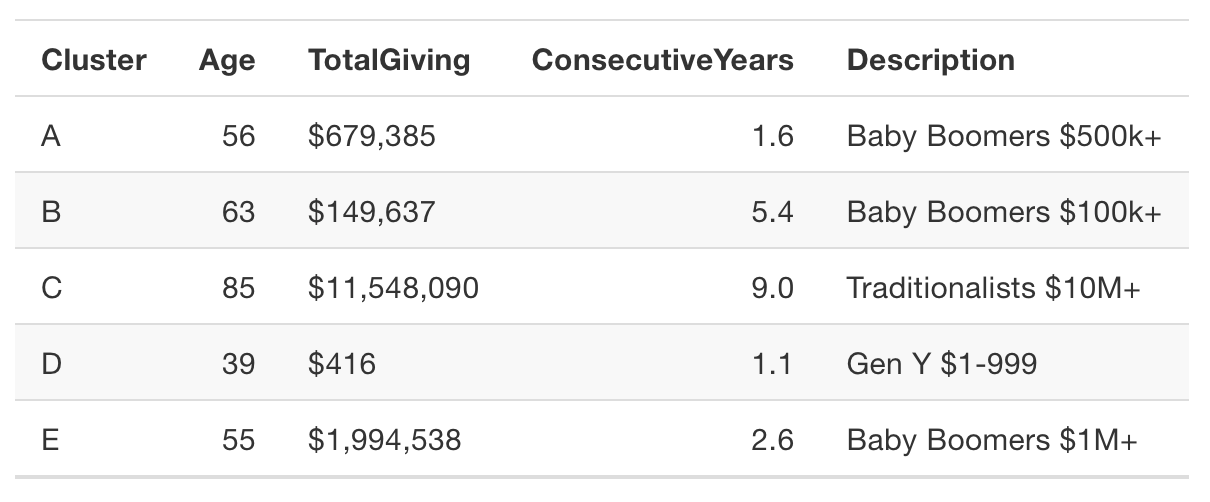
FIGURE 12.3: K-means donor cluster analysis
Cluster analysis, which is a type of unsupervised learning, is an effective method to explore patterns and create segments (groupings) within your data.
predict(model, newdata, type) function call to export probabilities instead of class predictions (that is, donor versus non-donor). Add these class probabilities into your kmeans cluster analysis, explore various segment or grouping sizes (k), and plot any useful cluster patterns (such as giving likelihood versus AGE, giving likelihood versus HAS_INVOLVEMENT_IND) with your own insight descriptions.
12.3.2 Association Rule Mining
Association rule mining (also known as market basket analysis) helps us find items that are frequently purchased together or, in our case, combinations of designations that are frequently donated together(that is, which designation combinations are common among our donors). This can help us uncover new prospective donors who have not given to a particular designation but have given somewhere else. For example, let’s say that through our analysis we know that donors who give to athletics also give to undergraduate scholarships. What if we looked at all the donors to athletics who have not given to undergraduate scholarships yet? That population might be very likely to give to scholarships, which we identified for future marketing segmentation and appeals.
To perform market basket analysis, we need the transaction history of donors and the designations they supported. You may run the following SQL code to get this data.
SELECT DONOR_ID, DESIGNATION
FROM GIVING_HISTORYFor our recipe, we will use a synthetic data set. Let’s load the arules and arulesViz libraries we need for this exercise(Hahsler et al. 2011; Hahsler 2017).
library(arules)
library(arulesViz)Let’s load the sample data set using the read.transactions function from the arules library.
giving_transactions <- read.transactions(
"data/SampleGivingTransactionsAssocRules.csv",
format = "single",
sep = ",",
cols = c("donor_id", "designation"))Let’s see what this object formatting looks like when using the arules library.
inspect(giving_transactions[1:3])
#> items transactionID
#> [1] {Arts,
#> JARVIS Studies Center,
#> Law,
#> Libraries,
#> Music,
#> Rugby,
#> Social Welfare,
#> Student Life} 1
#> [2] {Athletics,
#> English,
#> Innovation Fund,
#> JARVIS Studies Center,
#> Law,
#> Libraries,
#> Music,
#> Scholarships,
#> Student Life,
#> Superherology,
#> Theater} 10
#> [3] {Athletics,
#> JARVIS Studies Center,
#> Law,
#> Music,
#> Scholarships,
#> Student Life} 100Let’s also add a summary of the transactions.
summary(giving_transactions)
#> transactions as itemMatrix in sparse format with
#> 1000 rows (elements/itemsets/transactions) and
#> 20 columns (items) and a density of 0.431
#>
#> most frequent items:
#> Music Student Life Law
#> 881 856 854
#> Libraries Scholarships (Other)
#> 760 669 4610
#>
#> element (itemset/transaction) length distribution:
#> sizes
#> 1 2 3 4 5 6 7 8 9 10 11 12
#> 7 6 13 45 52 90 114 151 135 140 108 71
#> 13 14 15 16 17
#> 39 15 11 2 1
#>
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 1.00 7.00 9.00 8.63 10.00 17.00
#>
#> includes extended item information - examples:
#> labels
#> 1 Arts
#> 2 Arts Museum
#> 3 Athletics
#>
#> includes extended transaction information - examples:
#> transactionID
#> 1 1
#> 2 10
#> 3 100Let’s build the association rules using the apriori function. Two important parameters for association rule mining are confidence and support. The confidence parameter controls the reliability of each rule. If it is set too low, we will get many rules with designations that don’t show up frequently together. If it is set too high, we will get only a few rules, but with designation-combinations that are given too frequently. Similarly, support is a measure of the frequency of designations in the rules.
giving_rules <- apriori(giving_transactions,
parameter = list(supp = 0.5,
conf = 0.9))
#> Apriori
#>
#> Parameter specification:
#> confidence minval smax arem aval
#> 0.9 0.1 1 none FALSE
#> originalSupport maxtime support minlen maxlen
#> TRUE 5 0.5 1 10
#> target ext
#> rules FALSE
#>
#> Algorithmic control:
#> filter tree heap memopt load sort verbose
#> 0.1 TRUE TRUE FALSE TRUE 2 TRUE
#>
#> Absolute minimum support count: 500
#>
#> set item appearances ...[0 item(s)] done [0.00s].
#> set transactions ...[20 item(s), 1000 transaction(s)] done [0.00s].
#> sorting and recoding items ... [8 item(s)] done [0.00s].
#> creating transaction tree ... done [0.00s].
#> checking subsets of size 1 2 3 4 done [0.00s].
#> writing ... [21 rule(s)] done [0.00s].
#> creating S4 object ... done [0.00s].Let’s sort the rules by confidence (that is, the rules with highest confidence will rise to the top).
giving_rules <- sort(giving_rules,
by = 'confidence',
decreasing = TRUE)Let’s see a few of these rules.
inspect(giving_rules[1:10])
#> lhs rhs support confidence lift count
#> [1] {Law,
#> Rugby} => {Music} 0.509 0.931 1.06 509
#> [2] {Rugby,
#> Student Life} => {Music} 0.522 0.929 1.05 522
#> [3] {Law,
#> Libraries,
#> Student Life} => {Music} 0.566 0.928 1.05 566
#> [4] {Libraries,
#> Music,
#> Student Life} => {Law} 0.566 0.920 1.08 566
#> [5] {Law,
#> Scholarships} => {Music} 0.550 0.918 1.04 550
#> [6] {Scholarships,
#> Student Life} => {Music} 0.537 0.918 1.04 537
#> [7] {Law,
#> Rugby} => {Student Life} 0.502 0.918 1.07 502
#> [8] {Law,
#> Libraries} => {Music} 0.619 0.917 1.04 619
#> [9] {Rugby} => {Music} 0.574 0.915 1.04 574
#> [10] {Libraries,
#> Student Life} => {Music} 0.615 0.915 1.04 615It seems like the donors who gave to Law and to Rugby also gave to Music. Similarly, the donors who gave to Rugby and Student Life also gave to Music. The lhs and rhs indicate which designations of giving go together. For example, the first rule is {Law,Rugby} => {Music}, which means donors who gave to Law and Rugby also gave to Music.
We can see rules specific to certain designations. For example, if we want to see all the rules where the rhs is Law (that is, where else the donors give to, we can use the subset function).
inspect(subset(giving_rules, (rhs %in% 'Law')))
#> lhs rhs support confidence lift count
#> [1] {Libraries,
#> Music,
#> Student Life} => {Law} 0.566 0.920 1.08 566
#> [2] {Scholarships,
#> Student Life} => {Law} 0.533 0.911 1.07 533
#> [3] {Libraries,
#> Student Life} => {Law} 0.610 0.908 1.06 610
#> [4] {Music,
#> Scholarships} => {Law} 0.550 0.908 1.06 550
#> [5] {Libraries,
#> Music} => {Law} 0.619 0.905 1.06 619The arulesViz library offers multiple ways to visualize the generated rules. Let’s see a few of them, but first let’s store the top 10 rules for plotting.
top_10_giving_rules <- head(giving_rules, 10)Let’s plot a scatterplot between confidence and support.
plot(giving_rules) It looks like that there are many rules with high confidence and support between 0.5 and 0.6. These rules also have high lift.
It looks like that there are many rules with high confidence and support between 0.5 and 0.6. These rules also have high lift.
Next, let’s view the rules as a parallel coordinate plot.
plot(top_10_giving_rules,
method = "paracoord",
control = list(reorder = TRUE))
This parallel coordinate plot shows that the donors often give to Student Life, Music, Libraries, and Law designations. Also, many donors who give to other areas also give to Music.
Let’s see a grouped plot. This plot shows the rules in a matrix form.
plot(top_10_giving_rules, method = "grouped")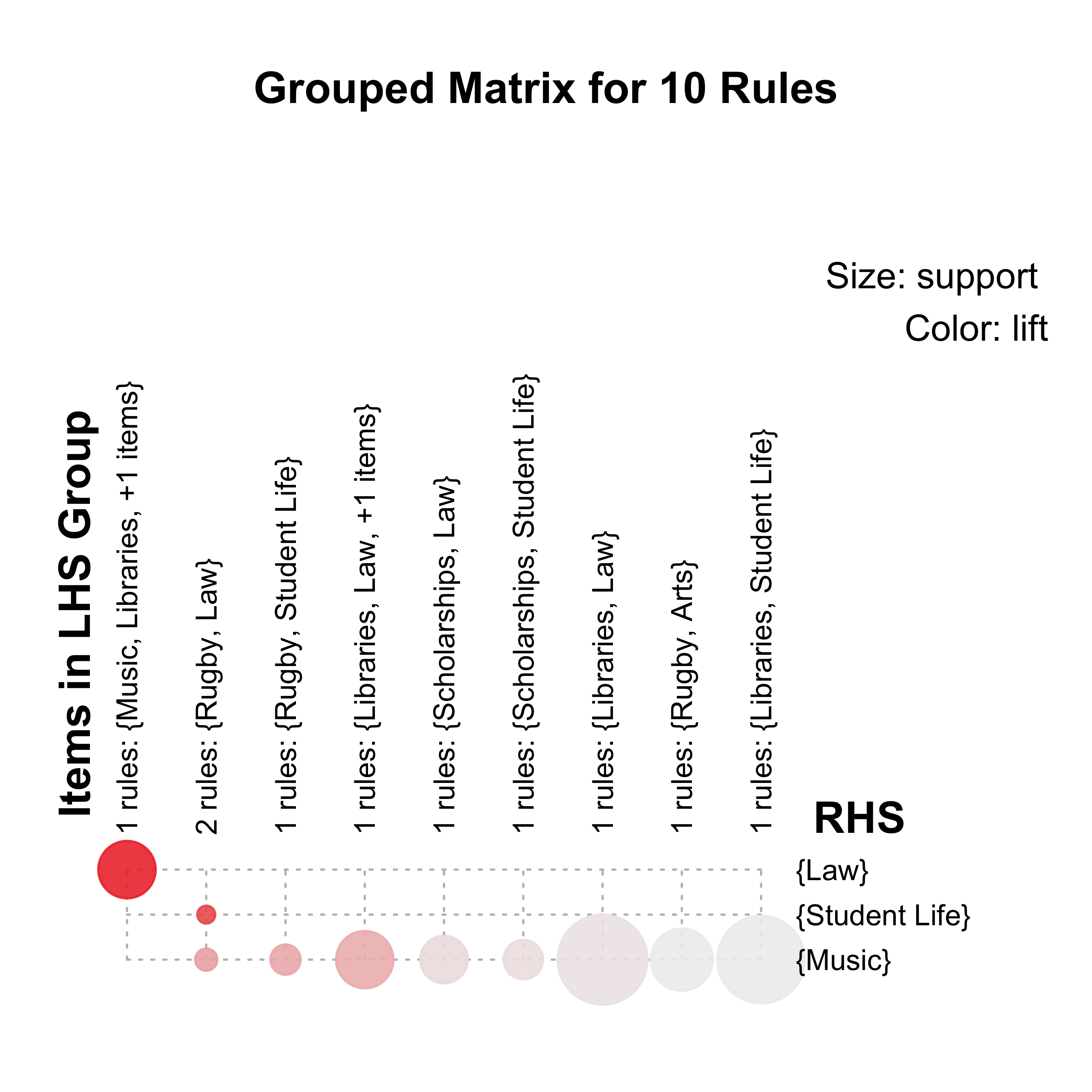
It is easy to see from this plot that many donors who gave to other areas also gave to Music and that the rule {Libraries,Music,Student Life} => {Law} has the highest support and lift.
Finally, let’s see these rules in network graph format, a topic we will explore in detail in the Social Network Analysis chapter.
plot(top_10_giving_rules, method = "graph")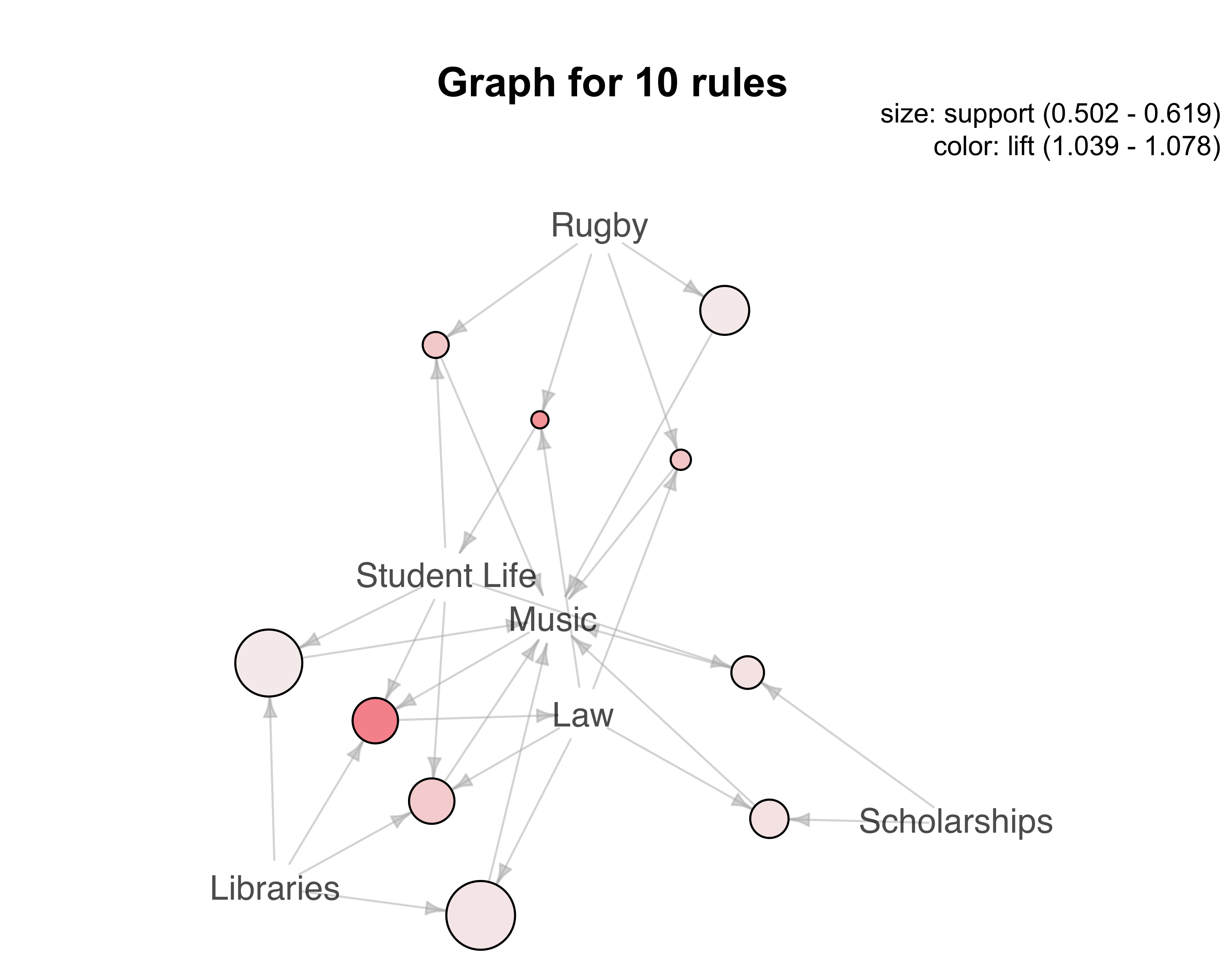 This network graph provides a different way of looking at the rules, specifically the relationships among various designations.
This network graph provides a different way of looking at the rules, specifically the relationships among various designations.
12.4 Model Diagnostics
As you build and refine your models, you will have to find practical ways to diagnose, tune and improve your model’s performance. While detailed model diagnostics and performance tuning is beyond the scope (and purpose) of this book, it is important that you understand the fundamental concepts of bias, variance, and the bias-variance tradeoff.
Bias refers to errors due to incorrect model assumptions with your machine learning algorithm, such as independence of variables. Models with high bias tend to miss (underfit) important relations between the model’s input and output variables. Variance refers to model errors generated due to sensitivity to small fluctuations in your training data. Models with high variance are prone to over-fitting to “noise” (random error) in your training data.
Bias and variance both contribute to your model’s total error, which can be represented by the following formula:
Total Error = Bias2 + Variance + Irreducible Error
Irreducible Error (also known as inherent uncertainty) is error usually associated with randomness or innate variability in a system. Since we cannot change irreducible error, we need to focus our attention on the bias and variance in our models. To deal with bias and variance, you can experiment with tuning your model’s parameters, revisit your model specification, seek data for missing features, and/or try different machine learning algorithms.
Ultimately, to get good predictions, you will need to strike a balance between bias and variance that minimizes the overall total error of your model. To learn more about bias and variance, we recommend reading this bias-variance tradeoff article as a starting point.
To learn more about using the mlr package to evaluate model performance and parameter tuning, you can check out these Evaluating Learner Performance, Resampling, and Tuning Hyperparameters articles.
12.5 Summary
In this chapter, we introduced and explored fundamental machine learning concepts and practical recipes using R to build fundraising solutions.
After reading this chapter, you should be able to recognize a variety of supervised and unsupervised machine learning algorithms as well as some commonly used R packages to implement solutions for both classification (categorical) and regression (numerical) tasks using your institutional data. It is important that you run these recipes on your data to cement the learning and get sensible results. Whether you are seeking to forecast numerical values or predict categorical outcomes, R, in combination with its vast array of machine learning packages, is a powerful, versatile, and evolving tool that you can use to build data analytics solutions and add value to your organization.
For a more extensive exploration of various machine learning methods and detailed coverage of model performance and evaluation techniques, you can check out the books Machine Learning with R by Lantz (2013) and Practical Machine Learning Tools and Techniques by Witten and Frank (2005).
If you’re enjoying this book, consider sharing it with your network by running
source("http://arn.la/shareds4fr")in yourRconsole.— Ashutosh and Rodger
References
Box, G. E. P. 1976. “Science and Statistics.” Journal of the American Statistical Association 71: 791–99. http://www.tandfonline.com/doi/abs/10.1080/01621459.1976.10480949.
Burnham, D. R., K. P.; Anderson. 2002. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. Springer-Verlag.
Warnes, Gregory R., Ben Bolker, Thomas Lumley, Randall C Johnson, and Randall C. Johnson. 2015. Gmodels: Various R Programming Tools for Model Fitting. https://CRAN.R-project.org/package=gmodels.
Ripley, Brian. 2015. Class: Functions for Classification. https://CRAN.R-project.org/package=class.
Jed Wing, Max Kuhn. Contributions from, Steve Weston, Andre Williams, Chris Keefer, Allan Engelhardt, Tony Cooper, Zachary Mayer, et al. 2017. Caret: Classification and Regression Training. https://CRAN.R-project.org/package=caret.
Meyer, David, Evgenia Dimitriadou, Kurt Hornik, Andreas Weingessel, and Friedrich Leisch. 2017. E1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), Tu Wien. https://CRAN.R-project.org/package=e1071.
Therneau, Terry, Beth Atkinson, and Brian Ripley. 2017. Rpart: Recursive Partitioning and Regression Trees. https://CRAN.R-project.org/package=rpart.
Milborrow, Stephen. 2017. Rpart.plot: Plot ’Rpart’ Models: An Enhanced Version of ’Plot.rpart’. https://CRAN.R-project.org/package=rpart.plot.
Hothorn, Torsten, Kurt Hornik, Carolin Strobl, and Achim Zeileis. 2017. Party: A Laboratory for Recursive Partytioning. https://CRAN.R-project.org/package=party.
Kuhn, Max, and Ross Quinlan. 2017. C50: C5.0 Decision Trees and Rule-Based Models. https://CRAN.R-project.org/package=C50.
Liaw, Andy, Matthew Wiener, and others. 2002. “Classification and Regression by randomForest.” R News 2 (3): 18–22.
Breiman, Leo, Adele Cutler, Andy Liaw, and Matthew Wiener. 2015. RandomForest: Breiman and Cutler’s Random Forests for Classification and Regression. https://CRAN.R-project.org/package=randomForest.
Chen, Tianqi, Tong He, Michael Benesty, Vadim Khotilovich, and Yuan Tang. 2017. Xgboost: Extreme Gradient Boosting. https://CRAN.R-project.org/package=xgboost.
Holte, Robert C. 1993. “Very Simple Classification Rules Perform Well on Most Commonly Used Datasets.” Machine Learning 11 (1). Springer: 63–90.
Witten, Ian H., and Eibe Frank. 2005. Data Mining: Practical Machine Learning Tools and Techniques. 2nd ed. San Francisco: Morgan Kaufmann.
Hornik, Kurt, Christian Buchta, and Achim Zeileis. 2009. “Open-Source Machine Learning: R Meets Weka.” Computational Statistics 24 (2): 225–32.
Cohen, W. W. 1995. “Fast Effective Rule Induction.” Proceedings of the 12th International Conference on Machine Learning, 115–23. http://citeseer.ist.psu.edu/cohen95fast.html.
Nandeshwar, Ashutosh R. 2006. “Models for Calculating Confidence Intervals for Neural Networks.” Master’s thesis, West Virginia University Libraries.
Abadi, Martín, Ashish Agarwal, Paul Barham, Eugene Brevdo, Zhifeng Chen, Craig Citro, Greg S Corrado, et al. 2016. “Tensorflow: Large-Scale Machine Learning on Heterogeneous Distributed Systems.” arXiv Preprint arXiv:1603.04467.
Cheng, Heng-Tze, Lichan Hong, Mustafa Ispir, Clemens Mewald, Zakaria Haque, Illia Polosukhin, Georgios Roumpos, et al. 2017. “TensorFlow Estimators: Managing Simplicity Vs. Flexibility in High-Level Machine Learning Frameworks.” In Proceedings of the 23rd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, 1763–71. New York, NY, USA: ACM. http://doi.acm.org/10.1145/3097983.3098171.
Tang, Yuan, JJ Allaire, RStudio, Kevin Ushey, Daniel Falbel, and Google Inc. 2017. Tfestimators: High-Level Estimator Interface to Tensorflow in R. https://github.com/rstudio/tfestimators.
Hahsler, Michael, Sudheer Chelluboina, Kurt Hornik, and Christian Buchta. 2011. “The Arules R-Package Ecosystem: Analyzing Interesting Patterns from Large Transaction Datasets.” Journal of Machine Learning Research 12: 1977–81. http://jmlr.csail.mit.edu/papers/v12/hahsler11a.html.
Hahsler, Michael. 2017. ArulesViz: Visualizing Association Rules and Frequent Itemsets. https://CRAN.R-project.org/package=arulesViz.
Lantz, Brett. 2013. Machine Learning with R. Packt.